- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurement of Total Phosphorus and Polyphosphate in Chlamydomonas reinhardtii
(*contributed equally to this work) Published: Vol 13, Iss 11, Jun 5, 2023 DOI: 10.21769/BioProtoc.4692 Views: 1509
Reviewed by: Ansul LokdarshiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
High-Performance Liquid Chromatography Quantification of Glyphosate, Aminomethylphosphonic Acid, and Ascorbate in Culture Medium and Microalgal Cells
Juan Manuel Ostera [...] Gabriela Malanga
Apr 5, 2025 1168 Views
Isolation of Mitochondria From Yeast to Estimate Mitochondrial Pools of Inorganic Phosphate
Swagata Adhikary [...] Sunil Laxman
Jul 5, 2025 1709 Views
Quantification of Protochlorophyllide (Pchlide) Content in Arabidopsis Seedlings Using a High-Performance Liquid Chromatography (HPLC) System
Fan Zhang [...] Liangsheng Wang
Jan 5, 2026 552 Views
Abstract
Phosphorus is an essential nutrient for plants. Green algae usually store excess P as polyphosphate (polyP) in the vacuoles. PolyP, a linear chain of three to hundreds of phosphate residues linked by phosphoanhydride bonds, is important for cell growth. Based on the previous method of polyP purification with silica gel columns (Werner et al., 2005; Canadell et al., 2016) in yeast cells, we developed a protocol to purify and determine the total P and polyP in Chlamydomonas reinhardtii by a quick, simplified, and quantitative method. We use hydrochloric acid or nitric acid to digest polyP or total P in dried cells and analyze P content using the malachite green colorimetric method. This method may be applied to other microalgae.
Keywords: Green algaeBackground
As an essential nutrient, phosphorus is required for the constitution of cellular components such as nucleic acids, phospholipids, and ATP. P is also required for the regulation of protein phosphorylation in plants. P is stored as inorganic phosphate (Pi) in the vacuoles of land plants and as inorganic polyphosphate (polyP) in chlorophyte algae. PolyP is a linear chain composed of several phosphate residues that are linked together by phosphoanhydride bonds. PolyP is an important metabolite and a signaling molecule acting as an energy source, a regulator of gene expression, a channel-forming component, and a storage form of Pi found in green plants (Wang et al., 2021).
Since the polyP is a mixture with different chain lengths, few analytical methods are currently available for polyP quantitation. In order to determine the content of polyP, it usually has to be degraded into Pi first, and then its content can be estimated by measuring the amount of Pi released. Two experimental methods are widely used for quantitatively estimating the amount of polyP in vitro (Aschar-Sobbi et al., 2008). The first one is based on the hydrolysis of polyP by hydrochloric acid and subsequent measurement of the Pi released. The second one is based on the activity of two enzymes, exopolyphosphatase and polyphosphate kinase, which can digest and convert the polyP into Pi for measurement. However, the expression and purification of these enzymes are complicated and time consuming (Christ et al., 2020).
Here, we describe an updated protocol for quantitative measurement of the polyP content in algae. We use silica gel columns to purify polyP and then convert it into Pi through digestion by hydrochloric acid. Although this method is not very accurate, because some short-chain-length polyP could not be completely extracted by the silica gel columns, it is economical and convenient. This protocol is relatively simple and time saving, leading to a fast determination of the polyP in Chlamydomonas, and should be considered suitable for polyP determination in other microalgae.
Materials and reagents
Centrifuge tubes (1.5, 2, 15, and 50 mL) (Sangon Biotech)
Sterile culture flasks (50 and 250 mL) (Sangon Biotech)
Pipettes (10, 20 and 200 μL, 1 and 5 mL) (Eppendorf)
96-well ELISA plate (Sangon Biotech, catalog number: F605031)
Petri dishes (Sangon Biotech, catalog number: F611003)
47 mm filter paper sheets (Whatman, catalog number: 1822047)
Spin Columns CA2 from TIANgel Midi Purification kit (TIANGEN, catalog number: DP209)
Hydrochloric acid (HCl)
Nitric acid (HNO3)
98% sulfuric acid (H2SO4)
Tris (C4H11NO3) (Sangon Biotech, catalog number: A100826)
Glacial acetic acid (CH3COOH) (Sangon Biotech, catalog number: A501931)
Ammonium chloride (NH4Cl) (Sangon Biotech, catalog number: A501569)
Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sangon Biotech, catalog number: A610329)
Calcium chloride dihydrate (CaCl2·2H2O) (Sangon Biotech, catalog number: A610050)
Dipotassium hydrogen phosphate (K2HPO4) (Sangon Biotech, catalog number: A610447)
Potassium phosphate monobasic (KH2PO4) (Sangon Biotech, catalog number: A600445)
EDTA disodium salt (C10H14N2O8Na2·2H2O) (Sangon Biotech, catalog number: A100105)
Zinc sulfate heptahydrate (ZnSO4·7H2O) (Sangon Biotech, catalog number: A602906)
Iron(II) sulfate heptahydrate (FeSO4·7H2O) (Sangon Biotech, catalog number: A600461)
Boric acid (H3BO3) (Sangon Biotech, catalog number: A100588)
Manganese (II) chloride tetrahydrate (MnCl2·4H2O) (Sangon Biotech, catalog number: A500331)
Cobalt chloride hexahydrate (CoCl2·6H2O) (Sigma-Aldrich: catalog number: 255599)
Copper(II) sulfate pentahydrate (CuSO4·5H2O) (Sangon Biotech, catalog number: A600063)
Ethanol absolute (Sangon Biotech, catalog number: A500737)
Neutral red (Sangon Biotech, catalog number: A600895)
1 M Tris-HCl (pH 7.5) (Sangon Biotech, catalog number: A610283)
Sodium iodide (NaI) (Sangon Biotech, catalog number: A610283)
Ammonium heptamolybdate tetrahydrate [(NH4)6Mo7O24·4H2O] (Sangon Biotech, catalog number: A600067)
Malachite green oxalate salt (Sangon Biotech, catalog number: A620330)
TAP medium (see Recipes)
40× TAP salt mixture (see Recipes)
Phosphate solution (50 μg/mL) (see Recipes)
Hutner's trace elements (see Recipes)
TAP solid medium (see Recipes)
1 M sulfuric acid (H2SO4) (see Recipes)
2 M NaOH (see Recipes)
70% ethanol (see Recipes)
0.1% neutral red solution (w/v) (see Recipes)
1 M Tris-HCl (pH 7.5) supplemented to 6% (v/v) with 0.1% neutral red solution (w/v) (see Recipes)
6 M NaI (see Recipes)
Wash buffer (see Recipes)
2 M hydrochloric acid (HCl) (see Recipes)
Phosphate (Pi) solution (10 μg/mL) (see Recipes)
28 mM ammonium heptamolybdate in 2.1 M H2SO4 (see Recipes)
0.76 mM malachite green in 0.35% polyvinyl alcohol (see Recipes)
Equipment
Spectrophotometer (Shimadzu, model: UVmini-1240)
Centrifuge (Thermo Scientific, catalog number: 75002411)
Hitachi Himac CT15RE tabletop high-speed microcentrifuge
Hitachi CR21N high-speed refrigerated centrifuge
Vortex (IKA, model: IKA MS 3 basic)
Thermal cycler (Thermo Scientific, catalog number: 4484073)
Microplate reader (Tecan, model: Infinite® F50 microplate reader)
Vacuum pump (JinTeng, model: GM-0.5B)
Analytical balance (Sangon Biotech, catalog number: G001142)
Air dry oven (Sangon Biotech, catalog number: G003409)
Clean bench (AIRTECH, catalog number: SW-CJ-2F)
Fume hood (ALPHA, VAV controller)
Nikon Digital Camera D7200
Software
Microsoft Excel (Microsoft, Inc.)
Microsoft PowerPoint (Microsoft, Inc.)
Prism GraphPad (Dotmatic, Inc.)
Procedure
In this section, we describe the procedure to measure polyP and total P in C. reinhardtii. The first step is to culture algae cells. Next, collect an appropriate number of algae cells to extract P (polyP or total P) and then digest polyP or total P into Pi by hydrochloric acid or nitric acid from dried cells. To quantify the Pi released, the malachite green colorimetric method is used.
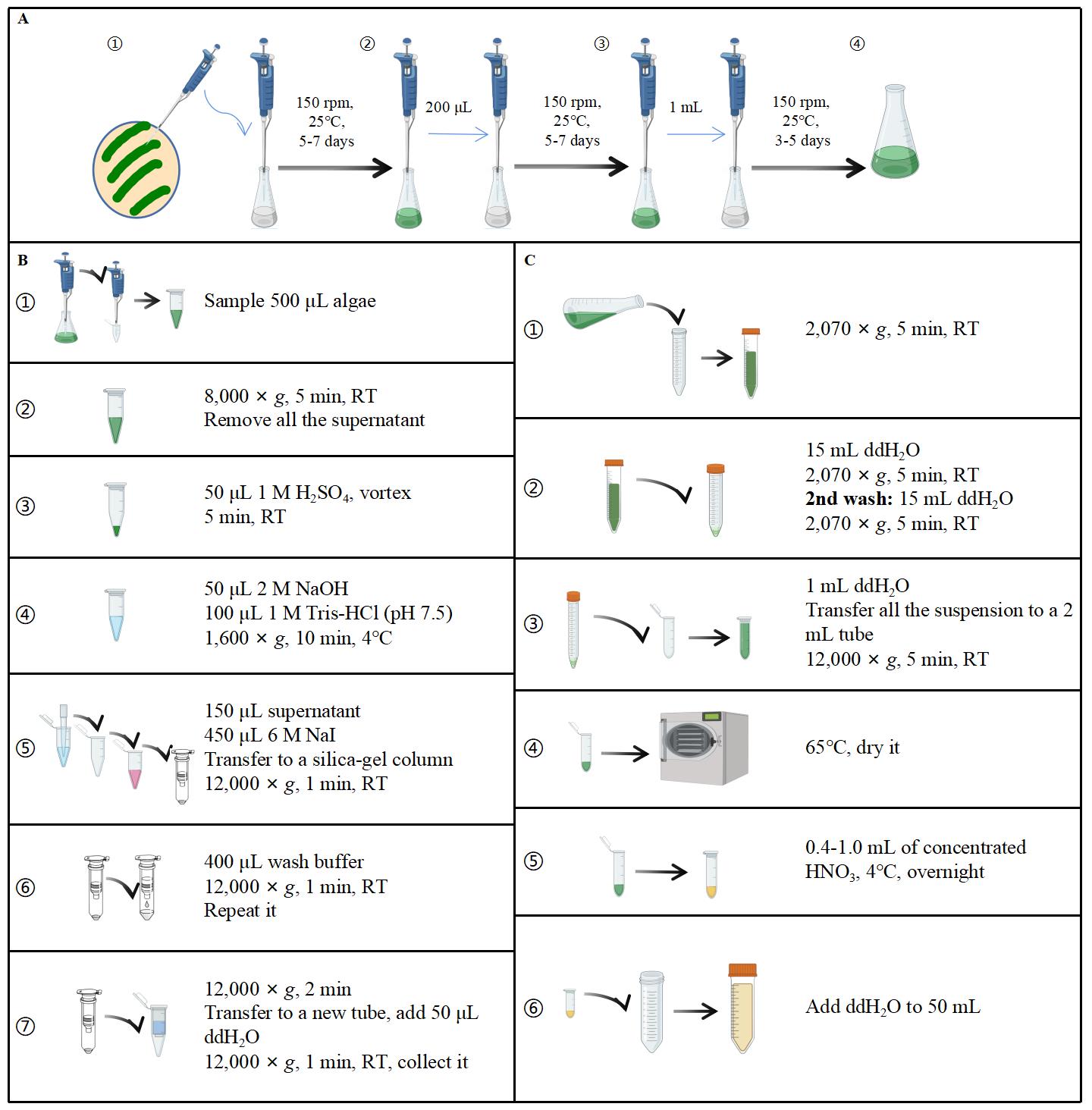
Figure 1. Basic workflow of cell culture and P measurement. A. Procedure for algae cell culture. B and C show the procedures for polyP and total P measurement, respectively. Drawn and composed using Microsoft PowerPoint.
Culture methods
Protocols adapted from Wang et al. (2021).
Cell culture is grown using 20 mL of TAP medium in a 50 mL flask. Stationary-phase cell culture with a volume of 200 μL is transferred into 20 mL of fresh medium every week. The culture growth conditions are adjusted to a speed of 150 rpm shaking at 25 °C using a rocking incubator, and the culture itself is illuminated continuously with white fluorescent light (150 μmol photons m-2 s-1). A cell density of approximately 1 × 107 cells (3–5 days after transfer, late linear phase) is used for the experiment.
The following provides specific steps:
Pick wildtype (CC-4533) and mutant (Crptc1) or other desired strain(s) from a fresh plate with a sterile toothpick or pipette tip and inoculate with 20 mL of liquid medium (TAP medium or appropriate selection medium) using a 50 mL flask. Rotate at 150 rpm and 25 °C for 5–7 days under continuous white light. This primary culture is grown to a stationary phase.
Inoculate cultures into a 50 mL flask with 20 mL of TAP medium and adjust inoculum size appropriately to make the OD750 of inoculated solution approximately 0.2–0.3. Cultures will reach the stable period (~1 × 107 cells) again after 5–7 days. The OD750 should be measured every 12 h to count the number of algae cells.
Transfer 1 mL of the secondary algae cells into a 250 mL flask containing 100 mL of TAP medium and incubate for 3–5 days.
After three times of uniformization, different algae strains reach the same or similar growth status.
Optional:
Determine the OD750 every 12 h, calculate the regression equation, and draw the growth curve.
Note: The growth phase can be determined by measuring cell density and drawing the growth curve.
Extraction of P
This section contains two parts. The first one is the purification and degradation of polyP and the second one is the digestion of total P in algae with nitric acid. After purification and degradation, polyP and total phosphorus content is converted into Pi released and can be measured using the malachite green colorimetric method.
Purification and degradation of polyP
Protocol adapted from Werner et al. (2005), Canadell et al. (2016), and Wang et al. (2021).
Determine the OD750 of the algae using a spectrophotometer and record it.
Add 500 μL of algae (record the exact weight) into a 1.5 mL centrifuge tube. After centrifugation at 8,000× g for 5 min at room temperature, remove all the supernatant by pipetting and harvest the algae cells.
Note: The sample can be directly purified or stored at -20 °C. For more accurate measurements of dry weight, 5 mL of algae is filtered on 47 mm filter paper sheets using a vacuum pump, and then put in a drying oven for several hours (Figure 1). The weight of the 47 mm filter paper sheet should be measured using an analytical balance before and after sample loading.
Add 50 μL of 1 M H2SO4 (see Recipes) into the algae, mix by pipetting or vortex to homogenate, and then place the tube at room temperature for 5 min.
Neutralize the suspension with 50 μL of 2 M NaOH (see Recipes) and 100 μL of 1 M Tris-HCl (pH 7.5) supplemented to 6% (v/v) with 0.1% neutral red solution (see Recipes). Gently mix the tubes upside down four times.
Note: The pH indicator helps to assure that the pH is around 7 (the sample color becomes orange red), which is a critical step for the reproducibility of polyP extraction. Samples too acidic (pink) or too basic (yellow) have to be corrected to the appropriate pH (orange red) by the addition of 2 M NaOH or 1 M H2SO4, respectively.
Centrifuge at 1,600× g for 10 min at 4 °C.
Drain 150 μL of supernatant with a pipette into a new 1.5 mL centrifuge tube containing 450 μL of 6 M NaI (freshly prepared; see Recipes).
Mix by pipetting 3–5 times and then transfer to a silica gel column (Spin Columns CA2 from TIANgel Midi Purification kit). Centrifuge the column at 12,000× g for 1 min at room temperature.
Discard the flowthrough and place the column back into the collection tube. Add 400 μL of wash buffer (see Recipes) into the column and centrifuge the column at 12,000× g for 1 min at room temperature.
Repeat step B1h.
Discard the flowthrough, place the column back into the collection tube, and centrifuge at 12,000× g for 2 min at room temperature to remove residual wash buffer.
Transfer the column into a clean 1.5 mL centrifuge tube and place the column with the cap open for 2–5 min to dry the membrane.
Add 50 μL of ddH2O to the center of the membrane, incubate at room temperature for 2 min, and then centrifuge at 12,000× g for 2 min at room temperature.
Note: The eluted polyP can be directly degraded; however, leaving it at room temperature for a longer time should be avoided. For long-term storage, it can be kept at -20 °C.
Pipette 10 μL of eluted polyP and add 10 μL of 2 M HCl (see Recipes), mix thoroughly, and centrifuge for several seconds.
Incubate at 95 °C for 30 min in the thermal cycler and then transfer onto the ice.
The degraded polyP sample can be directly measured with the malachite green colorimetric method or stored at -20 °C.
Determination of total P
Mark a 2 mL centrifuge tube and weigh it.
Collect 10–15 mL of cells into a 15 mL centrifuge tube and centrifuge at 2,070× g for 5 min at room temperature.
Discard the supernatant, wash the algae cells with ddH2O, and centrifuge at 2,070× g for 5 min at room temperature.
Repeat step B2c.
Discard the supernatant, suspend the algae cells with 1 mL of ddH2O, and transfer the entire suspension to the 2 mL centrifuge tube.
Centrifuge at 12,000× g for 5 min at room temperature and pipette all the supernatant carefully in case of loss of algae samples.
Dry and weigh the tube with algae samples.
Add 0.4–1.0 mL of concentrated HNO3 and incubate overnight at 4 °C until the solution is clean.
Transfer the solution into a 50 mL centrifuge tube, wash the inside of the 2 mL centrifuge tube with ddH2O, transfer all the wash water into the 50 mL centrifuge tube, and dilute with ddH2O to 50 mL.
This sample can then be analyzed for P concentration determination using the malachite green colorimetric method.
Quantification of P with the malachite green colorimetric method
Drawing of the standard curve (Table 1)
Table 1. Standard curve
1 2 3 4 5 6 7 Concentration μg/mL 0 0.25 0.5 0.75 1 1.5 2 Pi solution (10 μg/mL) μL 0 2.5 5 7.5 10 15 20 ddH2O μL 100 97.5 95 92.5 90 85 80 According to the Pi concentration gradient in the table, add these substances in order:
Add Pi solution (10 μg/mL) (see Recipes) and the corresponding volume of ddH2O.
Add 86 μL of 28 mM ammonium heptamolybdate in 2.1 M H2SO4 (see Recipes).
Add 64 μL of 0.76 mM malachite green in 0.35% polyvinyl alcohol (see Recipes).
Leave the 96-well ELISA plate at room temperature for 30 min and measure the OD595 with a microplate reader.
Note: Do not let the reaction proceed for more than 1 h as it can cause the appearance of small precipitates in highly concentrated Pi samples and polyP degradation in background samples, thus interfering with the correct Pi measure.
Draw the standard curve by taking the volume of Pi solution (10 μg/mL) as the x-axis and the OD595 as the y-axis. Then, the P concentration of the samples can be calculated according to the standard curve.
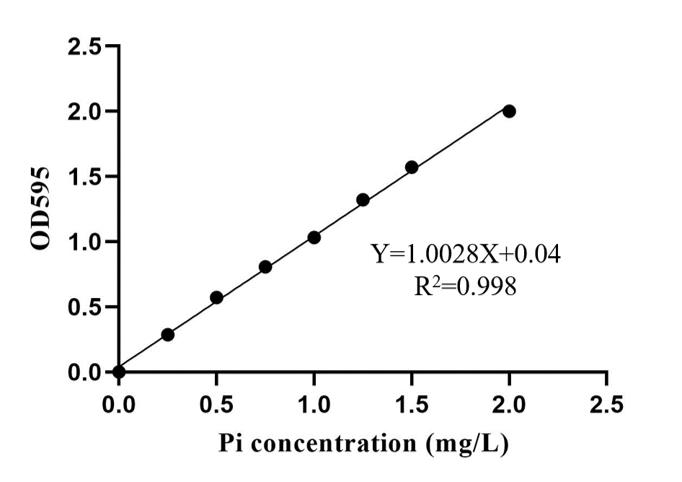
Figure 2. Standard curve. Pi concentration as x-axis and the OD595 as the y-axis. Drawn using Microsoft Excel and Prism GraphPad.Determination of P in samples
Use a 96-well plate for the reaction and measurement of polyP. To quantify released Pi, add to each well 10 μL of polyP after degradation, 90 μL of ddH2O, 86 μL of 28 mM ammonium heptamolybdate in 2.1 M H2SO4 (see Recipes), and 64 μL of 0.76 mM malachite green in 0.35% polyvinyl alcohol (see Recipes), in this order. Mix the well by pipetting gently.
The blank control consists of 100 μL of ddH2O, 86 μL of 28 mM ammonium heptamolybdate in 2.1 M H2SO4, and 64 μL of 0.76 mM malachite green in 0.35% polyvinyl alcohol.
Leave the 96-well plate at room temperature for 30 min and measure the OD595 with a microplate reader.
Results
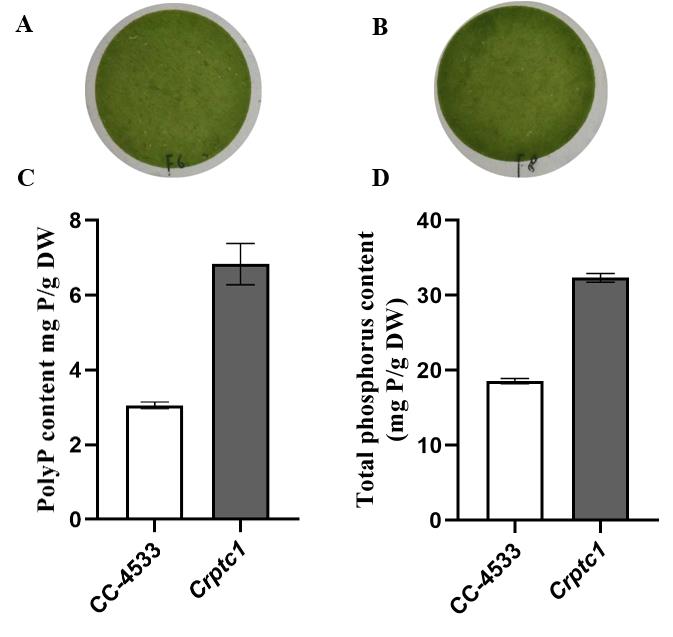
Figure 3. The content of PolyP and total P in wildtype strain CC-4533 and the Crptc1 mutant. (A) and (B) show filter paper sheets of 5 mL cells through a vacuum pump. The weight of algae cells was calculated using the dry weight difference of filter paper before and after filtration. PolyP content (C) and total P content (D) of the wildtype strain CC-4533 and the Crptc1 mutant. CrPTC1 is a vacuole efflux phosphate transporter in algae and catalyzes Pi transport out of acidocalcisomes. The Crptc1 mutant over-accumulates total P and polyP (Wang et al., 2021). Data is presented as the mean ± SD from five independent experiments. Pictures were taken using a Nikon Digital Camera D7200. C and D were drawn using Microsoft Excel and Prism GraphPad.
Recipes
TAP medium
Tris 2.42 g
40× TAP salt mixture 25 mL (Recipe 2)
Phosphate solution (50 μg/mL) 0.4 mL (Recipe 3)
Hutner’s trace elements 1 mL (Recipe 4)
Glacial acetic acid 1 mL
Adjust to pH 7.0 with HCl
Add ddH2O to 1 L and autoclave at 121 °C for 20 min
40× TAP salt mixture
NH4Cl 15 g
MgSO4·7H2O 4 g
CaCl2·2H2O 2 g
Add ddH2O to 1 L
Phosphate solution (50 μg/mL)
K2HPO4 28.8 g
KH2PO4 14.4 g
Add ddH2O to 100 mL
Hutner’s trace elements
EDTA disodium salt 50 g
ZnSO4·7H2O 22 g
FeSO4·7H2O 4.99 g
H3BO3 11.4 g
MnCl2·4H2O 5.06 g
CoCl2·6H2O 1.61 g
CuSO4·5H2O 1.57 g
(NH4)6Mo7O24·4H2O 1.10 g
Add ddH2O to 1 L
TAP solid medium
TAP medium (Recipe 1) with 1% (w/v) agar
Autoclave at 121 °C for 20 min
1 M sulfuric acid (H2SO4)
Mix 5.45 mL of 98% sulfuric acid with 94.55 mL of ddH2O and store at room temperature
2 M NaOH
Mix 8 g of NaOH with 80 mL of ddH2O
Add ddH2O to 100 mL and store at room temperature
70% ethanol
Mix 70 mL of ethanol absolute with 30 mL of ddH2O
0.1% neutral red solution (w/v)
Mix 0.1 g of neutral red with 100 mL of 70% ethanol (Recipe 8) and store at room temperature
1 M Tris-HCl (pH 7.5) supplemented to 6% (v/v) with 0.1% neutral red solution (w/v)
Mix 12.1 g of Tris with 70 mL of ddH2O
Add 6 mL of 0.1% neutral red solution (w/v) (Recipe 9)
Adjust pH to 7.5 with HCl
Add ddH2O to 100 mL and store at room temperature
6 M NaI
Mix 8.9934 g of NaI with ddH2O up to 10 mL
The solution has to be freshly prepared each time and kept in darkness
Wash buffer
10 mM Tris-HCl buffer (pH 7.5), 50% ethanol, 1 mM EDTA, and 100 mM NaCl
Mix 5 mL of 1 M Tris-HCl (pH 7.5), 250 mL of 100% ethanol, 1 mL of 0.5 M EDTA, 10 mL of 5 M NaCl, and 200 mL of ddH2O
Adjust pH to 7.5 with HCl
Add ddH2O to 500 mL and store at room temperature
2 M HCl
Mix 16.67 mL of 37% HCl with 83.33 mL of ddH2O and store at room temperature
Phosphate (Pi) solution (10 μg/mL)
Mix 50 μL of Phosphate solution (50 μg/mL; Recipe 3) and 200 μL of ddH2O
28 mM ammonium heptamolybdate in 2.1 M H2SO4
Mix 3.46 g of ammonium heptamolybdate tetrahydrate with 80 mL of ddH2O
Add 11.2 mL of 98% H2SO4
Add ddH2O to 100 mL and store at room temperature
0.76 mM malachite green in 0.35% polyvinyl alcohol
Mix 350 mg of polyvinyl alcohol in 100 mL of ddH2O at 80 °C and stir vigorously on a magnetic stirrer until all polyvinyl alcohol dissolves completely
Add 35 mg of malachite green oxalate salt
Add ddH2O to 100 mL and store at room temperature
Acknowledgments
The protocol for algae culture was adapted from Wang et al. (2021). The protocol for polyP purification was modified from Werner et al. (2005) and Canadell et al. (2016). This work was supported by the National Natural Science Foundation of China (32130096, 32202593, and 32102478) and the National Key Research and Development Program of China (2021YFF1000404). K.Y. was supported by the Innovation Program of the Chinese Academy of Agricultural Sciences. X.J. and L.W. were supported by the China Postdoctoral Science Foundation (2021M693447 to X.J.; 2021M693449 and 2022T150707 to L.W.).
Competing interests
No conflict of interest or competing interests declared.
References
- Wang, L., Jia, X., Zhang, Y., Xu, L., Menand, B., Zhao, H., Zeng, H., Dolan, L., Zhu, Y. and Yi, K. (2021). Loss of two families of SPX domain-containing proteins required for vacuolar polyphosphate accumulation coincides with the transition to phosphate storage in green plants. Mol Plant 14(5): 838-846.
- Aschar-Sobbi, R., Abramov, A. Y., Diao, C., Kargacin, M. E., Kargacin, G. J., French, R. J. and Pavlov, E. (2008). High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J Fluoresc 18(5): 859-866.
- Christ, J. J., Willbold, S. and Blank, L. M. (2020). Methods for the analysis of polyphosphate in the life sciences. Anal Chem 92(6): 4167-4176.
- Werner, T. P., Amrhein, N. and Freimoser, F. M. (2005). Novel method for the quantification of inorganic polyphosphate (iPoP) in Saccharomyces cerevisiae shows dependence of iPoP content on the growth phase. Arch Microbiol 184(2): 129-136.
- Canadell, D., Bru, S., Clotet, J. and Ariño, J. (2016). Extraction and quantification of polyphosphate in the budding yeast Saccharomyces cerevisiae. Bio-protocol 6(14): e1874.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Yang, Y., Ren, S., Jia, X., Zeng, H., Wang, L., Zhu, Y. and Yi, K. (2023). Measurement of Total Phosphorus and Polyphosphate in Chlamydomonas reinhardtii. Bio-protocol 13(11): e4692. DOI: 10.21769/BioProtoc.4692.
Category
Plant Science > Plant biochemistry > Other compound
Biochemistry > Other compound > Polyphosphate
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









