- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Reconstitution of Phase-separated p62 Bodies on the Arp2/3-derived Actin Network
Published: Vol 13, Iss 8, Apr 20, 2023 DOI: 10.21769/BioProtoc.4656 Views: 2234
Reviewed by: Oneil Girish BhalalaZhongmin LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
An Efficient Method for Immortalizing Mouse Embryonic Fibroblasts by CRISPR-mediated Deletion of the Tp53 Gene
Srisathya Srinivasan and Hsin-Yi Henry Ho
Jan 20, 2025 2740 Views
Puromycin Proximity Ligation Assay (Puro-PLA) to Assess Local Translation in Axons From Human Neurons
Raffaella De Pace [...] Saikat Ghosh
Mar 5, 2025 3282 Views
Assay for Site-Specific Homologous Recombination Activity in Adherent Cells, Suspension Cells, and Tumor Tissues
Yuki Yoshino [...] Natsuko Chiba
Apr 5, 2025 2426 Views
Abstract
In cells, p62/SQSTM1 undergoes liquid–liquid phase separation (LLPS) with poly-ubiquitin chains to form p62 bodies that work as a hub for various cellular events, including selective autophagy. Cytoskeleton components such as Arp2/3-derived branched actin network and motor protein myosin 1D have been shown to actively participate in the formation of phase-separated p62 bodies. Here, we describe a detailed protocol on the purification of p62 and other proteins, the assembly of the branched actin network, and the reconstitution of p62 bodies along with cytoskeletal structures in vitro. This cell-free reconstitution of p62 bodies vividly mimics the phenomenon in which low concentrations of protein in vivo rely on cytoskeleton dynamics to increase the local concentration to reach the threshold for phase separation. This protocol provides an easily implemented and typical model system to study cytoskeleton-involved protein phase separation.
Keywords: p62Background
Liquid–liquid phase separation (LLPS) plays critical roles in orchestrating biomolecular function and assembly of membrane-less organelles during diverse cellular processes (Banani et al., 2017). How LLPS is spatiotemporally regulated in the context of complicated cellular structures and components remains largely unknown. We recently revealed that cytoskeleton, as a solid phase, can actively participate in the phase separation of liquid-phase protein condensates, by using p62 bodies as a model system (Feng et al., 2022). P62/SQSTM1, a selective autophagy receptor, mediates LLPS of poly-ubiquitinated proteins into large condensates, which are also known as p62 bodies (Sun et al., 2018; Zaffagnini et al., 2018). The phase-separated p62 bodies can act as nucleation site for autophagosome formation, being then degraded by autophagy. Failure or inappropriate formation of p62 bodies is associated with pathological development of neurodegenerative diseases and Paget's disease, a chronic bone disease characterized by abnormal and excessive bone loss (Wong and Cuervo, 2010; Zaffagnini et al., 2018).
Despite p62 protein and polyubiquitin chain being shown to form large condensates in vitro (Sun et al., 2018), we reveal that the dendritic branched actin network is required for large p62 body formation in vivo. By establishing an in vitro reconstitution assay, we further demonstrate that such cytoskeleton components can greatly lower the protein concentration threshold during p62 body formation (Feng et al., 2022). The implication of cytoskeleton in LLPS represents an important and probably general way to achieve precise spatiotemporal regulation of biomolecular phase separation. Therefore, establishing an in vitro phase separation assay on cytoskeleton network is necessary and critical for these research fields; however, such protocol has not been clearly described before. Here, we present a detailed protocol to in vitro reconstitute p62 bodies along with branched actin network components in Lab Tek Chambered cover glasses. Actin polymerization is initiated by adding actin monomer, Arp2/3 complex, capping protein (CapZ), and PWCA [a domain consisting of a conserved C-terminal proline-rich region and WASP-homology 2 peptides plus a connector and acidic segments, which promotes Arp2/3-mediated actin polymerization (Campellone et al., 2008)]. Motor protein Myo1D is also added to facilitate phase separation of p62 and poly-ubiquitin (Ub8). The in vitro–reconstituted p62 bodies can be visualized by total internal reflection fluorescence (TIRF) and scanning electronic microscopy. This protocol provides an instruction on how to set up an in vitro phase separation assay involving cytoskeletal components.
Materials and Reagents
HEK293F cells (ATCC, catalog number: CRL-1573)
Sf9 cells (ATCC, catalog number: CRL-1711)
BL21 (DE3) E. coli cells (TransGen Biotech, catalog number: CD601-02)
Lab Tek Chambered cover glass (Thermo Fisher Scientific, catalog number: 150682)
SMM 293-TII expression medium (Sino Biological, catalog number: M293TII)
Sf-900 II SFM medium (Thermo Fisher Scientific, catalog number: 10902096)
Polyethyleneimines (PEI) (Polysciences, catalog number: 23966-2)
Amylose beads (New England Biolabs, catalog number: E8035S)
LB broth powder (Sangon Biotech, catalog number: A507002)
Isopropyl-beta-D-thiogalactopyranoside (IPTG) solution (Sangon Biotech, catalog number: B541007)
Tris (Sigma-Aldrich, catalog number: 77-86-1)
Phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: 329-98-6)
Acetic acid (Sangon Biotech, catalog number: A501931)
Ammonium acetate (Sangon Biotech, catalog number: A600032)
Phosphate buffered saline (PBS) (HyClone, catalog number:SH80255.01)
HEPES (Sigma-Aldrich, catalog number: 7365-45-9)
Imidazole (Sigma-Aldrich, catalog number: 288-32-4)
Glycerol (Sinopharm, catalog number: 56-81-5)
DTT (dithiothreitol) (Inaclo, catalog number: 0281)
Maltose (Sigma-Aldrich, catalog number: 69-79-4)
Rosetta (DE3) pLysS-competent E. coli cells (TIANGEN, catalog number: CB108-02)
Strep-Tactin resin (IBA Lifesciences, catalog number: 2-1201-002)
D-desthiobiotin (Sigma-Aldrich, catalog number: D1411)
HisPur(tm) Ni-NTA Resin (Thermo Fisher Scientific, catalog number: 88222)
MBP Trap HP column (GE Healthcare, catalog number: 28-9187-78)
TEV protease (Solarbio, catalog number: P2060)
Sodium chloride (NaCl) (Sangon Biotech, catalog number: A610476)
PIPES (Sangon Biotech, catalog number: A620434)
Potassium chloride (KCl) (Sangon Biotech, catalog number: A610440)
MgCl2 (Sangon Biotech, catalog number: B601193)
EGTA (MedChemExpress, catalog number: HY-D0861)
Ethylenediaminetetraacetic acid (EDTA) (MedChemExpress, catalog number: HY-Y0682)
Adenosine 5'-triphosphate (ATP) (MedChemExpress, catalog number: HY-B2176)
MOPS (MedChemExpress, catalog number: HY-D0859)
Triton X-100 (Solarbio, catalog number: T8200)
Protease inhibitor cocktails (MedChemExpress, catalog number: HY-K0012)
Glucose oxidase (Solarbio, catalog number: G8030)
Catalase (Solarbio, catalog number: C8070)
Bovine serum albumin (BSA) (Solarbio, catalog number: A8010)
Methyl cellulose (MedChemExpress, catalog number: HY-125861)
Sucrose (Solarbio, catalog number: S8271)
ATP-Mg (Sigma Aldrich, catalog number: A9187)
Buffer A (see Recipes)
Buffer B (see Recipes)
Buffer C (see Recipes)
Lysis buffer (see Recipes)
Basic buffer (see Recipes)
Loading buffer (see Recipes)
High-salt buffer (see Recipes)
Wash buffer (see Recipes)
TIRF buffer (see Recipes)
Elution buffer (see Recipes)
Storage buffer (see Recipes)
Equipment
Nickel column (Bio-Rad, catalog number: 12009287)
Mono Q 5/50 GL column (GE Healthcare, catalog number: 17-5166-01)
HiTrap Q HP (GE Healthcare, catalog number: 17-1154-01)
Superdex200 Increase 10/300 GL (GE Healthcare, catalog number: 28990944)
Ultrafiltration discs (Sigma-Aldrich, catalog number: PLGC07610)
Allegra X-14 centrifuge, type 45 Ti rotor manual, JS-4.750 swinging-bucket rotor and buckets, 4 × 50 mL, 9 × 15 mL adapters (Beckman Coulter, model: Allegra X14, catalog number: A99464)
Water bath at 37 °C (Thermo Fisher Scientific, model: TSCIP19)
Incubator at 37 °C with 5% CO2 (Thermo Fisher Scientific, model: HERAcell 150i)
Confocal fluorescence microscopy (NIKON, A1 HD25)
Nikon SIM/STORM (TIRF)
Field emission scanning electron microscopy (FE-SEM) (FEI Quanta 200)
Critical Point Dryer (Shianjia Biotechnology, model: SCD-350M)
Software
NIS-Elements software (Laboratory Imaging)
ImageJ
Procedure
Purification of Arp2/3 complex
Construction of expression vector: Sub-clone the full-length open reading frames of seven subunits of human Arp2/3-ArpC1a-ArpC5 complex into pCAG vectors. To facilitate protein purification, add two strep-tags to the N-termini of ArpC1a (Doolittle et al., 2013a and 2013b).
Cell transfection: Grow HEK293F cells in SMM 293-TII expression medium to a density of 1.5–1.8 million cells per milliliter. Transiently transfect the plasmids to HEK293F cells using PEI. Mix 0.25 mg of plasmids of each subunit of complex (1.75 mg in total) and 3 mg of PEI in 50 mL of fresh medium for 25 min, then add to 800 mL of culture. Culture the transfected cells at 37 °C supplemented with 5% CO2 in a shaker for 48–72 h.
Extraction and purification of target proteins:
Harvest cells by centrifugation and resuspend in ice-cold buffer A.
Solubilize cell membrane using 1% Triton X-100 for 1 h while rotating at 4 °C.
Centrifuge cell lysates at 20,000 × g for 1 h at 4 °C using a type 45 Ti rotor to remove cell debris.
Collect and load the supernatant onto Strep-Tactin resin.
Remove unbound proteins by extensive wash with buffer B.
Elute the bound proteins with buffer B supplemented with 10 mM D-desthiobiotin.
Further purify the target protein by Superdex200 Increase 10/300 GL column using buffer C.
Pool and concentrate peak fractions containing all seven subunits to 10 mg/mL.
Purification of PWCA
Construction of vector: Clone the PWCA domain of human WHAMM protein into a pCAG vector containing a N-terminal MBP-tag (Liu et al., 2017). The PWCA domain is expressed and purified from the HEK293F cell line.
When the culture reaches a density of 1.2–1.6 million cells per milliliter, transfect cells with 1 mg of plasmids using 2 mg of PEI.
Extraction and purification of target proteins:
After 48–72 h post transfection, harvest cells and lyse in ice-cold lysis buffer for 1 h while rotating at 4 °C.
Remove the insoluble material by centrifugation at 20,000 × g for 1 h at 4 °C.
Incubate the supernatant with amylose beads for 2 h while rotating at 4 °C.
Remove unbound proteins by extensive wash with the basic buffer and elute the bound proteins using the same buffer with 10 mM maltose.
Further purify the eluted proteins by anion exchange using a Mono Q 5/50 GL column.
Load the proteins onto the Mono Q column at 1 mL/min using the loading buffer as running buffer.
Use high-salt buffer for gradient elution.
Pool and load the fractions containing the PWCA domain onto a Superdex 75 10/300 GL column for final purification.
Purification of capping protein (CapZ)
Construction of vector: Clone the cDNA of capping protein (α1β2, Mus musculus) into a pET-3d vector containing a N-terminal 6×His-tag.
Transfer the expression vector into Rosetta (DE3) pLysS-competent E. coli cells via transformation for protein expression.
Extraction and purification of target protein:
Grow bacteria in LB medium at 37 °C and induce protein expression at OD600 0.6–1.0 with 0.5 mM IPTG for 3 h.
Collect cell pellets by centrifugation and resuspend in ice-cold lysis buffer.
Lyse cells by ultrasonication; then, remove cell debris by centrifugation at 20,000 × g using the Type 45 Ti rotor at 4 °C for 1 h.
Collect the supernatant and load it onto Ni-NTA resin.
Wash the resin with wash buffer.
Elute the bound proteins with elution buffer.
Further purify the eluted proteins by anion exchange using a Mono Q 5/50 GL column.
Load the proteins onto the Mono Q column at 1 mL/min using the loading buffer as running buffer.
Use high-salt buffer for gradient elution.
Pool and load the fractions containing capping protein onto a Superdex 75 10/300 GL column for final purification.
Purification of myosin 1D
Grow Sf9 cells in sf-900 II SFM medium to a density of 2 × 106 cells/mL and incubate with virus harboring full-length Myo1D construct containing a N-terminal 6×His-tag.
After 60 h, collect cells, lyse by freeze-thaw cycles, and centrifuge at 4 °C.
Remove insoluble material by centrifugation at 20,000 × g for 30 min.
Incubate supernatants on a roller at 4 °C for 1 h with 1 mL of Ni-NTA resin and transfer into a column.
Wash the columns with wash buffer.
Elute myosin 1D with elution buffer.
The protein concentration of myosin 1D is determined by OD280.
Concentrate the protein by ultrafiltration discs (10 KD).
Store the protein in storage buffer.
Determine the purity of protein by Coomassie blue–stained SDS-poly-acrylamide gels.
Expression and purification of Ub8
Grow bacteria transformed with 6×His-Ub8 construct in LB medium at 37 °C and induce at OD600 ~0.8 with 0.5 mM IPTG at 18 °C for 16 h.
Re-suspend cells in 50 mM Tris (pH 8.0), 150 mM NaCl, and 2 mM phenylmethylsulphonyl fluoride (PMSF).
Transfer the cell suspension on ice and sonicate to disrupt the cells, followed by centrifugation at 47,850 × g for 1 h at 4 °C.
Isolate the proteins through a Ni-NTA column.
Further purify the proteins using a Hitrap Q HP column.
Acidify the solution of Ub8 using acetic acid.
Apply to a Hitrap SP HP column.
Elute with a linear gradient of NaCl (0–1 M) in 50 mM ammonium acetate pH 4.5.
Further purify Ub8 proteins by gel filtration. Equilibrate the gel filtration columns (Superdex 200 10/300) in 150 mM NaCl, 40 mM Tris-HCl (pH 7.4), 10% glycerol, and 1 mM DTT.
Flash-freeze Ub8 proteins and store at -80 °C.
Expression and purification of p62
Transfer the MBP-mCherry-p62 construct (His6-MBP-Tev-mCherry vector backbone) into BL21 (DE3) E. coli cells via transformation and spread to bacterial culture plate for overnight culture at 37 °C.
Pick up a single colony into a liquid LB medium (approximately 15 mL) and incubate at 37 °C in a bacterial shaker at 220 rpm for 14–18 h.
Inoculate 1% of the bacterial solution into 1 L liquid LB medium and expand the culture at 37 °C.
When OD600 reaches ~0.6, add 0.2 mM IPTG to induce protein expression at 16 °C overnight.
Collect bacteria at 1,800 × g for 15 min. Wash again with PBS.
Add 30 mL of lysis buffer, vortex, and shake to fully resuspend the bacteria.
Put the resuspended bacteria into a 50 mL glass beaker on ice.
Ultrasonic crush the bacteria for 10 min, at 60% intensity, sonication for 5 s, interval 3 s.
Centrifuge the sonication solution at 13,000 × g for 30 min at 4 °C.
Collect the supernatant and purify the proteins with MBP Trap HP column.
Use TEV protease to remove MBP tag from MBP-mCherry-p62 fusion protein (Raran-Kurussi et al., 2017). Add the equivalent amount of TEV protease to the standard reaction buffer (50 mM Tris-HCl pH 8.0, 0.5 mM EDTA, and 1 mM DTT) containing MBP-mCherry-p62 fusion protein and incubate overnight (4 °C) or 1–2 h (37 °C).
The resultant mCherry-p62 protein is used for in vitro phase separation assay.
In vitro reconstitution of p62 bodies on the Arp2/3-derived actin network (Figure 1)
Prepare actin monomers (1 μM, 50% Oregon-green labeled) as described in another protocol (Jiang and Huang, 2017). The growth of actin filaments can be observed under a Nikon microscope equipped with a 100× oil objective by the TIRF illumination.
To initiate branched actin polymerization, add 1 μM actin (50% Oregon-green labeled), 100 nM Arp2/3 complex, 600 nM CapZ, and 300 nM PWCA to a tube, then add TIRF buffer for a 50 μL mixed solution in total.
Carefully transfer 10 μL of mixed solution to the center of a Lab Tek Chambered cover glass.
In the focal plane of the chamber, robust branched actin polymerization can be observed by TIRF microscopy.
Add 10 μM Myo1D to the solution and continue TIRF imaging.
Add 0.5 μM mCherry-p62 and 3 μM Ub8 to the solution and continue TIRF imaging.
The reconstituted p62 body can also be examined by scanning electron microscopy. For the conventional fixation procedure, grow p62 bodies on coverslips, fix with 2.5% glutaraldehyde in PBS buffer for 2 h at room temperature, wash three times with PBS buffer, and then post-fix with 1% osmium for 20 min at room temperature. Then, dehydrate all samples with a graded series of ethanol (50%, 70%, 80%, 90%, 100%, and 100%) for 2 min each and dry with Critical Point Dryer using CO2. Coat the dried samples with an approximately 5 nm thick gold film by sputter coating before examination with focused ion beam scanning electron microscopy using a Helios detector at an acceleration voltage of 2.0 kV (Rong et al., 2012).
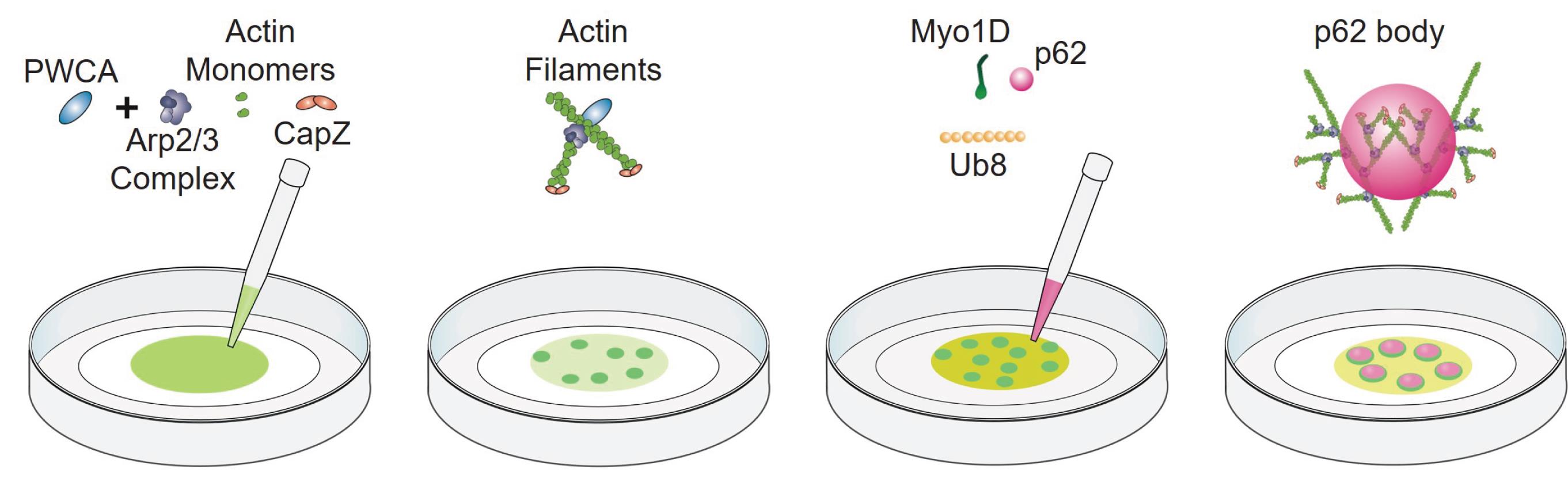
Figure 1. Schematic of the p62 body reconstitution assay. Molecules are not drawn to scale.
Data analysis
In this reconstitution assay, all components are derived from purified proteins. Oregon-green-labeled actin monomers can self-assemble into actin filaments (Figure 2, left panel). Addition of Arp2/3 complex, CapZ, and PWCA leads to the formation of branched actin network (Figure 2, right panel). Myo1D, as a motor protein, is not necessary for the branched actin network; however, its inclusion strengthens such branched actin structures. The scaffold proteins of p62 bodies (mCherry-p62 and Ub8) are then introduced on top of branched actin network to reconstitute cytoskeleton-associated p62 bodies, which can be observed by either TIRF microscopy or scanning electron microscopy (Figure 3 and Figure 4). Then, the NIS-Elements software is used for image measurement and co-location analysis. In such in vitro assay, phase separation of p62 bodies is greatly accelerated with lower concentration threshold of scaffold proteins in the presence of branched actin network than that using p62/Ub8 alone (Feng et al., 2022). Use ImageJ to process immunofluorescence images (rotating, cropping, and adjusting brightness and contrast when necessary).
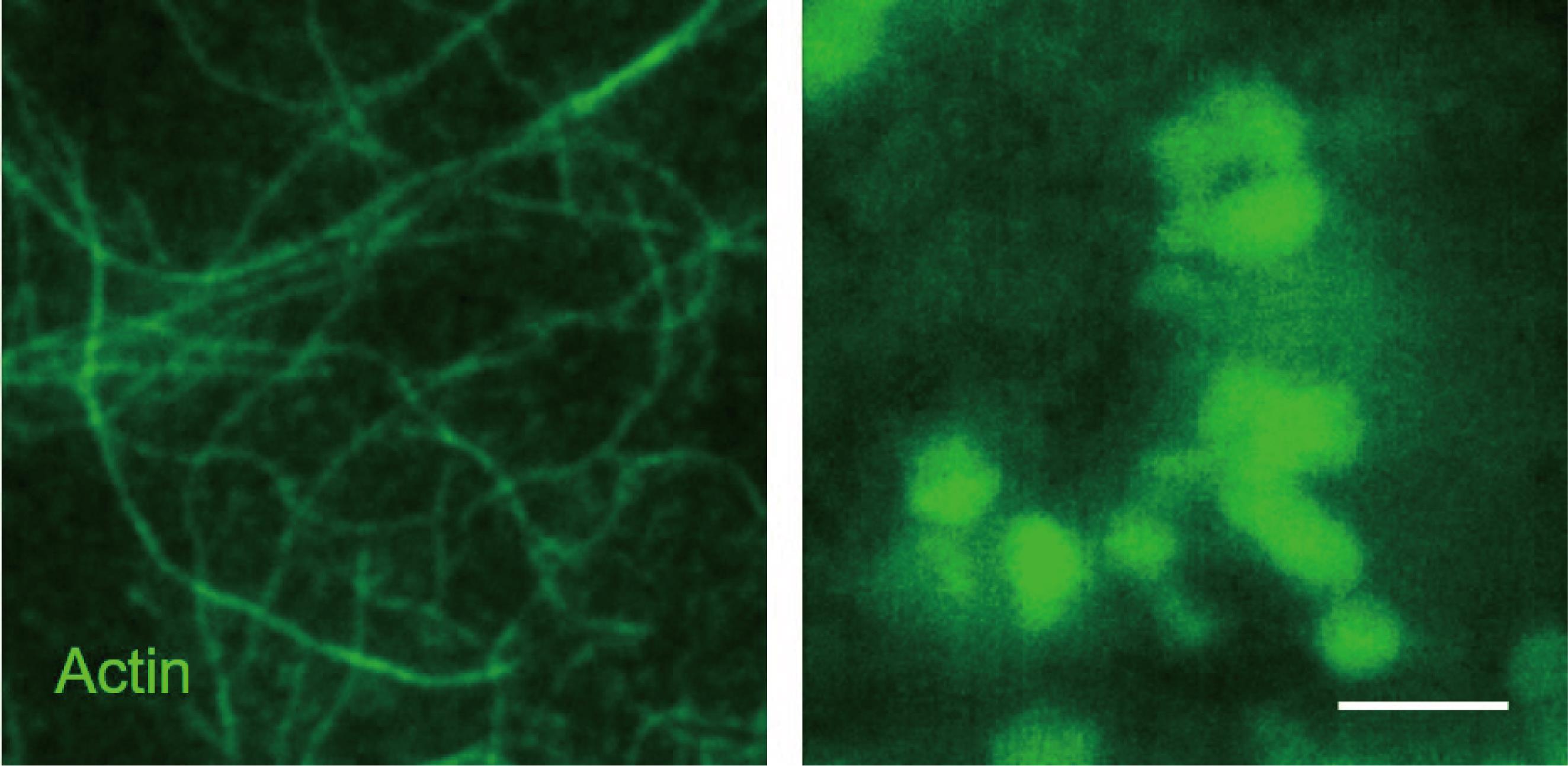
Figure 2. Total internal reflection fluorescence (TIRF) images showing the actin polymerization. Oregon-green-labeled actin filaments (left panel) and reconstituted branched actin network (right panel). Scale bar = 10 μm.
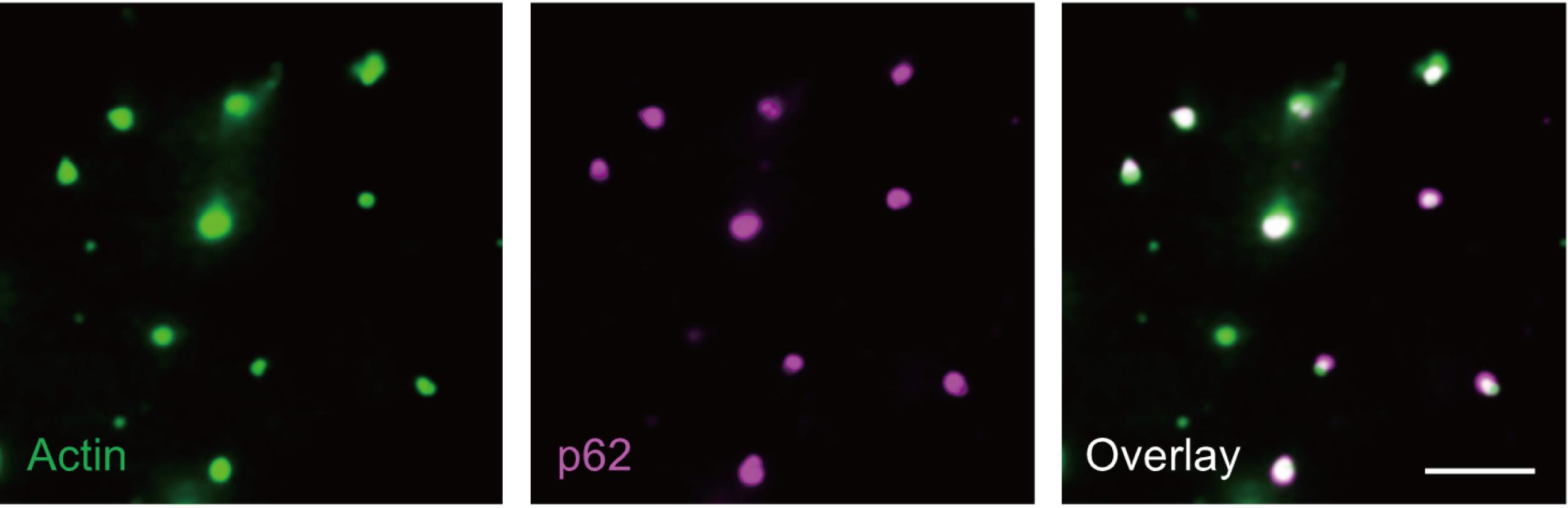
Figure 3. In vitro–reconstituted p62 bodies on branched actin network. Scale bar = 5 µm.
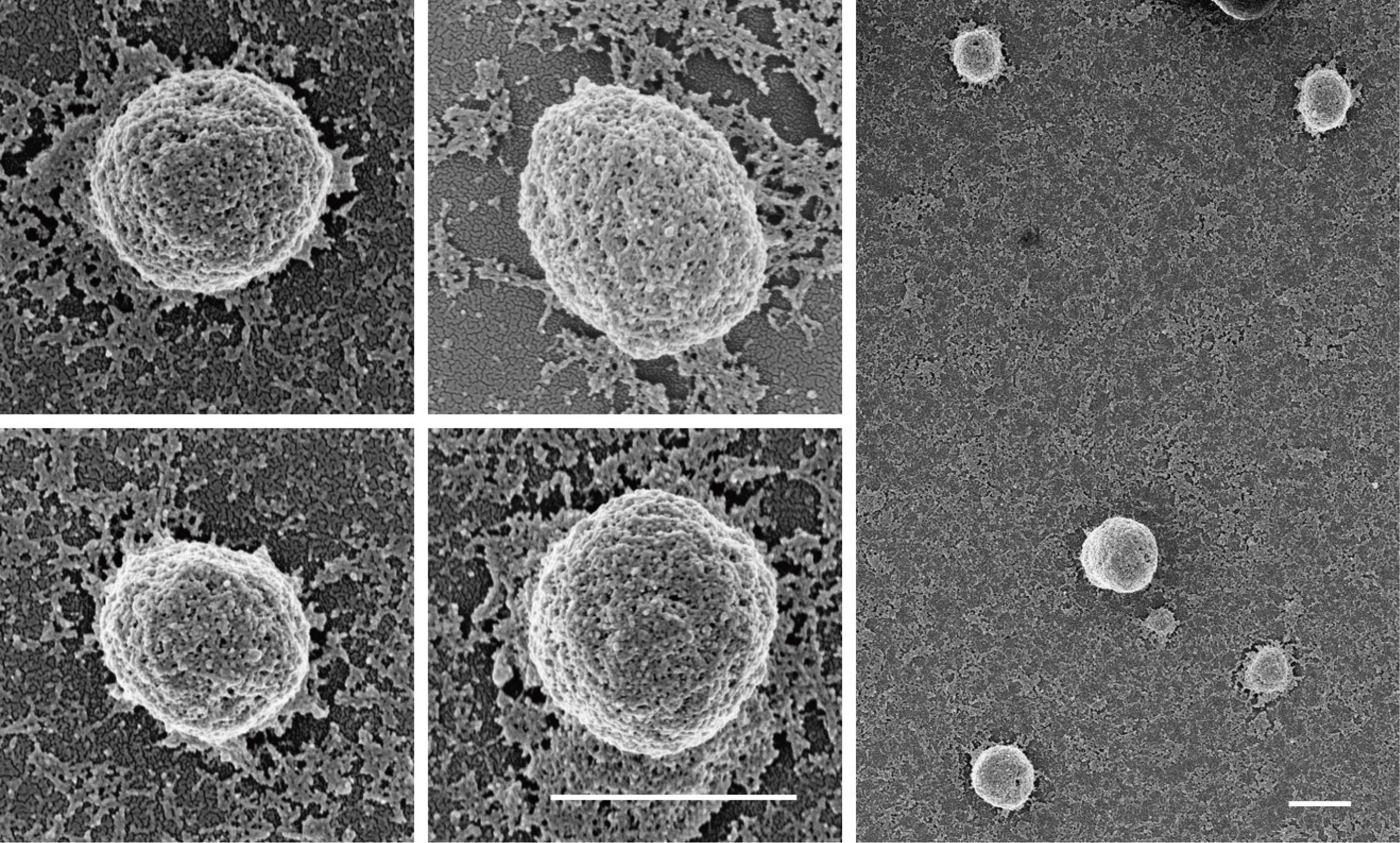
Figure 4. Morphology of reconstituted p62 bodies observed by FE-SEM. Field emission scanning electron microscopy (FE-SEM) image showing the structures of four reconstituted p62 bodies on the Arp2/3-derived actin network. Scale bar = 1 µm.
Notes
When mixing the components for branched actin polymerization, it is advised to firstly co-incubate PWCA and Arp2/3 complex for 5 min before mixing with actin and CapZ. Otherwise, the efficiency of branched actin polymerization will be decreased.
It is recommended to ultracentrifuge the actin monomers to remove the precipitate before use.
Recipes
Buffer A
20 mM HEPES pH 7.4
40 mM NaCl
1 mM DTT
Store at 4 °C, use within two weeks
Buffer B
20 mM PIPES pH 6.8
150 mM KCl
2 mM MgCl2
5 mM EGTA
1mM EDTA
0.5 mM DTT
0.2 mM ATP
5% glycerol
Store at 4 °C, use within two weeks
Buffer C
20 mM MOPS pH 7.0
100 mM KCl
2 mM MgCl2
5 mM EGTA
1 mM EDTA
0.5 mM DTT
0.2 mM ATP
5% glycerol
Store at 4 °C, use within two weeks
Lysis buffer
20 mM Tris pH 8.0
150 mM NaCl
1 mM DTT
1 mM EDTA
5% glycerol
1% Triton X-100
Protease inhibitor cocktails
Store at 4 °C, use within two weeks
Basic buffer
20 mM Tris pH 8.0
150 mM NaCl
1 mM DTT
1 mM EDTA
Store at 4 °C, use within two weeks
Loading buffer
20 mM Tris pH 8.0
80 mM NaCl
1 mM DTT
1 mM EDTA
Store at -20 °C
High-salt buffer
20 mM Tris pH 8.0
500 mM NaCl
1 mM DTT
1 mM EDTA
Store at 4 °C, use within two weeks
Wash buffer
20 mM HEPES pH 7.4
500 mM NaCl
1 mM DTT
5% glycerol
30 mM imidazole
Store at 4 °C, use within two weeks
TIRF buffer
1 mM MgCl2
50 mM KCl
1 mM EGTA
10 mM imidazole pH 8.0
0.2 mM ATP
100 mM DTT
100 μg/mL glucose oxidase
20 μg/mL catalase
0.2% BSA
0.5% methyl cellulose
Store at 4 °C, use within two weeks
Elution buffer
50 mM Tris pH 7.5
250 mM imidazole
300 mM NaCl
0.2 mM EGTA
Store at 4 °C, use within two weeks
Storage buffer
50 mM HEPES PH7.4
300 mM NaCl
50 μM ATP-Mg
10% sucrose
Store at 4 °C, use within two weeks
Acknowledgments
We thank Drs. Jiangfeng Shen, Xiaoyu Fu, Wanqing Du, Wenkang Zhao, Xuezhao Feng, Mingrui Ding, Jinpei Zhang and Daxiao Sun for their helpful efforts and suggestions on optimizing this protocol. This work was supported by the Ministry of Science and Technology of China (2017YFA0506300), and the National Natural Science Foundation of China (31771536 and 31860316).
Competing interests
The authors declare no competing interests.
References
- Banani, S. F., Lee, H. O., Hyman, A. A. and Rosen, M. K. (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18(5): 285-298.
- Campellone, K. G., Webb, N. J., Znameroski, E. A. and Welch, M. D. (2008). WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell 134(1): 148-161.
- Doolittle, L. K., Rosen, M. K. and Padrick, S. B. (2013a). Purification of native Arp2/3 complex from bovine thymus. Methods Mol Biol 1046: 231-250.
- Doolittle, L. K., Rosen, M. K. and Padrick, S. B. (2013b). Purification of Arp2/3 complex from Saccharomyces cerevisiae. Methods Mol Biol 1046: 251-271.
- Feng, X., Du, W., Ding, M., Zhao, W., Xirefu, X., Ma, M., Zhuang, Y., Fu, X., Shen, J., Zhang, J., et al. (2022). Myosin 1D and the branched actin network control the condensation of p62 bodies. Cell Res 32(7): 659-669.
- Jiang, Y. and Huang, S. (2017). Direct Visualization and Quantification of the Actin Nucleation andElongation Events in vitro by TIRF Microscopy. Bio Protoc 7(5): e2146.
- Liu, T., Dai, A., Cao, Y., Zhang, R., Dong, M. Q. and Wang, H. W. (2017). Structural Insights of WHAMM's Interaction with Microtubules by Cryo-EM. J Mol Biol 429(9): 1352-1363.
- Raran-Kurussi, S., Cherry, S., Zhang, D. and Waugh, D. S. (2017). Removal of Affinity Tags with TEV Protease. Methods Mol Biol 1586: 221-230.
- Rong, Y., Liu, M., Ma, L., Du, W., Zhang, H., Tian, Y., Cao, Z., Li, Y., Ren, H., Zhang, C., Li, L., Chen, S., Xi, J. and Yu, L. (2012). Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat Cell Biol 14(9): 924-934.
- Sun, D., Wu, R., Zheng, J., Li, P. and Yu, L. (2018). Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res 28(4): 405-415.
- Wong, E. and Cuervo, A. M. (2010). Autophagy gone awry in neurodegenerative diseases. Nat Neurosci 13(7): 805-811.
- Zaffagnini, G., Savova, A., Danieli, A., Romanov, J., Tremel, S., Ebner, M., Peterbauer, T., Sztacho, M., Trapannone, R., Tarafder, A. K., et al. (2018). p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J 37(5): e98308.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Liu, T., Xu, M. and Mi, N. (2023). In vitro Reconstitution of Phase-separated p62 Bodies on the Arp2/3-derived Actin Network. Bio-protocol 13(8): e4656. DOI: 10.21769/BioProtoc.4656.
Category
Biophysics > Microscopy
Biophysics > Biophotonics
Biochemistry > Protein > Self-assembly
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








