- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Quick DNA Extraction Method for High Throughput Screening in Gram-positive Bacteria
Published: Vol 13, Iss 8, Apr 20, 2023 DOI: 10.21769/BioProtoc.4653 Views: 2639
Reviewed by: Emilia KrypotouMarcelo S. da SilvaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation of Genomic DNA from Mycobacterium Species
Pawan Kumar [...] Sangeeta Bhaskar
Mar 5, 2016 17317 Views
Analysis of Replicative Intermediates of Adeno-associated Virus through Hirt Extraction and Southern Blotting
Martino Bardelli [...] Els Henckaerts
May 5, 2017 10565 Views
Implementation of Fusion Primer-Driven Racket PCR Protocol for Genome Walking
Yinwei Gu [...] Haixing Li
Dec 5, 2025 743 Views
Abstract
In this study, a sonication-based DNA extraction method was developed, in which the whole process can be finished within 10 min. This method is almost zero cost and time-saving, which is useful for high throughput screening, especially in the screening of mutants generated in random mutagenesis. This method is effective in genomic DNA extraction for PCR amplification in several Gram-positive bacteria, including Bacillus cereus, Bacillus thuringiensis, Bacillus subtilis, and Listeria monocytogenes.
Keywords: Gram-positive BacteriaBackground
Colony PCR is commonly used to verify the expected genome DNA in Escherichia coli, but it fails in Gram-positive bacteria such as Bacillus subtilis even when treated with chemical reagents (Packeiser et al., 2013; Azevedo et al., 2017). The reason colony PCR fails in these cases is the thick cell wall that exists in most Gram-positive bacteria, which impedes cell lysis and subsequent DNA release. Enzymes such as lysozyme, proteinase K, and other chemical reagents are used to lyse the Gram-positive bacterial cell wall (Mertens et al., 2014); however, these methods are costly, time consuming, and not eco-friendly for the DNA extraction of a huge quantity of samples. For example, it is very difficult to achieve a fast screen towards thousands of mutants generated in random mutagenesis. It is therefore necessary to set up a convenient and economical method.
Sonication is a common physical method employed to disrupt cells by transmitting ultrasonic energy to the liquid region (Taylor et al., 2001). Sonication-based DNA extraction is considered as an efficient and cost-effective method. In the intestinal microflora and compost microbes of fish, the ultrasonic lysis method combined with lysozyme was used to extract DNA (Yang et al., 2006; Han et al., 2018). Moreover, direct extraction of DNA from sputum clinical sample (containing diverse microbes) for real-time PCR proved to be convenient and suitable (Bello et al., 2020). In this study, a sonication-based DNA extraction method was developed, which could screen large samples in just one day. Despite not being as pure as the commercial kit, this method can be used for high-throughput screening purposes, which requires extracting great amounts of DNA, in Gram-positive bacteria such as Bacillus cereus, Bacillus thuringiensis, Bacillus subtilis, and Listeria monocytogenes.
Materials and Reagents
Materials
1.5 mL tube (Axygen, catalog number: MCT-150-C)
0.2 mL PCR tube (Axygen, catalog number: PCR-02-C-P)
10 and 100 μL pipettes (Eppendorf, catalog numbers: 3120000224 and 3120000828)
10 and 100 μL micropipettes (Axygen, catalog numbers: TGL-10FT-17-R and TF-100)
Petri dishes (Guangdong Huankai, catalog number: 20221700)
Inoculation loop (Guangdong Huankai, catalog number: 00416571)
Strains
Bacillus cereus ATCC 14579 (ATCC, catalog number: ATCC 14579)
Bacillus thuringiensis ATCC 10792 (ATCC, catalog number: ATCC 10792)
Bacillus subtilis ATCC 6633 (ATCC, catalog number: ATCC 6633)
Listeria monocytogenes ATCC 19115 (ATCC, catalog number: ATCC 19115)
Reagents
Nutrient agar plate (Guangdong Huankai, catalog number: 1024089)
Taq DNA polymerases (Thermo Fisher Scientific, catalog number: 10966018)
Marker D, 100–2,000 bp (Sangon Biotech, catalog number: B600335-0050)
GoldViewTM (Coolaber, catalog number: GD001-1mL*5)
50× TAE buffer (Sango Biotech, catalog number: B548101-0500)
Equipment
Incubator (SHANGHAI MINQUAN INSTRUMENT, model: LAB-0001-0004-SHMQ)
Benchtop centrifuge (Eppendorf, model: 5415D)
Ultrasonic cleaning machine (LANGEE, model: UC-9120)
Thermal cycler (Bio-Rad, model: C1000 Touch)
Electrophoresis imaging system (Bio-Rad, model: GelDoc XR Biorad)
Spectrophotometer (Thermo Fisher, model: NanoDropTM One Microvolume UV-Vis)
Autoclave (ZEALWAY, model: GR60DA)
Software
Image Lab (Bio-Rad)
Procedure
Cultivate the strains (listed in materials) conserved in glycerol at 37 °C, at 200 rpm overnight (for approximately 8–12 h).
Dip a small amount of the overnight culture using a 10 μL inoculation loop and streak lightly on the nutrient agar plate. Then, cultivate the plate at 37 °C overnight.
Suspend a single colony into 30 μL of ddH2O in a 1.5 mL tube.
Note: The size of the colonies of B. cereus ATCC 14579, B. thuringiensis ATCC 10792, B. subtilis ATCC 6633, and L. monocytogenes ATCC 19115 is recommended in Figure 1 and Table 1, and the quality of DNA is listed in Table 2.
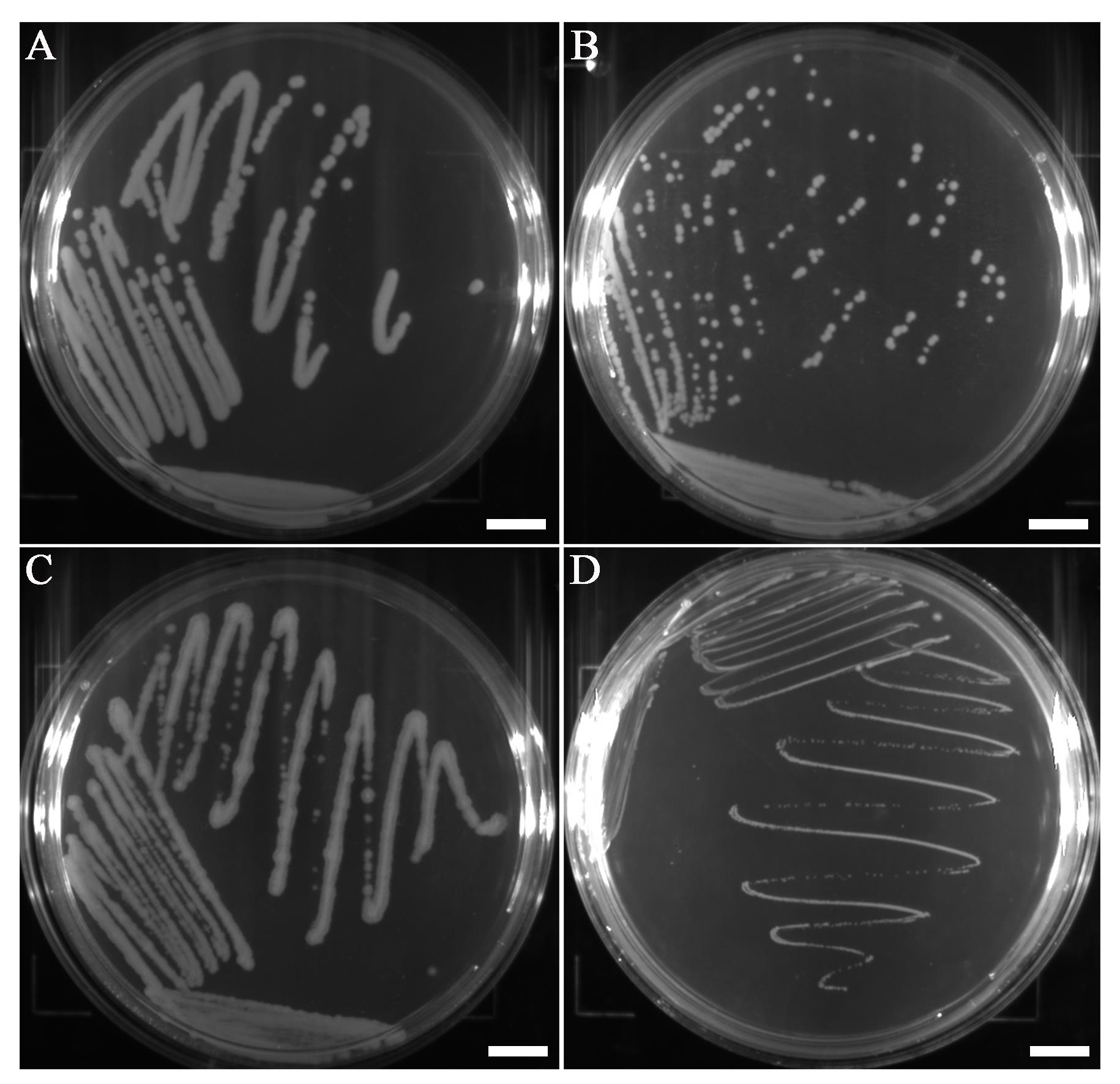
Figure 1. B. cereus (A), B. thuringiensis (B), B. subtilis (C), and L. monocytogenes (D) colonies formed on Petri dishes. Scale bars represent 10 mm.Table 1. Colony size of B. cereus, B. thuringiensis, B. subtilis, and L. monocytogenes
Microorganism Colony size B. cereus ATCC 14579 2–4 mm B. thuringiensis ATCC 10792 1–2 mm B. subtilis ATCC 6633 1–3 mm L. monocytogenes ATCC 19115 0.2–0.5 mm Treat the bacterial suspension by sonication at 40 kHz and 120 W at room temperature for 5 min and then centrifuge at 10,000 × g for 1 min at room temperature.
Note: No need to set pauses between the pulses.
Carefully transfer the upper aqueous phase to a new 1.5 mL tube without disturbing the pellet.
The DNA samples are tested using 1 μL of supernatant as DNA template by PCR. Primers for PCR are listed in Table 3. Taq DNA polymerases are added and parameters for PCR are as follows: initial denaturation: 95 °C, 3 min; denaturation: 95 °C, 30 s; annealing: 55 °C, 30 s; extension: 72 °C, 1 min; final extension: 72 °C, 10 min; number of cycles: 25×. After electrophoresis on 1.2% agarose gels (in 1× TAE diluent), all amplicons are visualized under UV light with GoldViewTM staining. The results are shown in Figure 2.
Note: The bands are brighter if you increase the PCR reaction cycles.
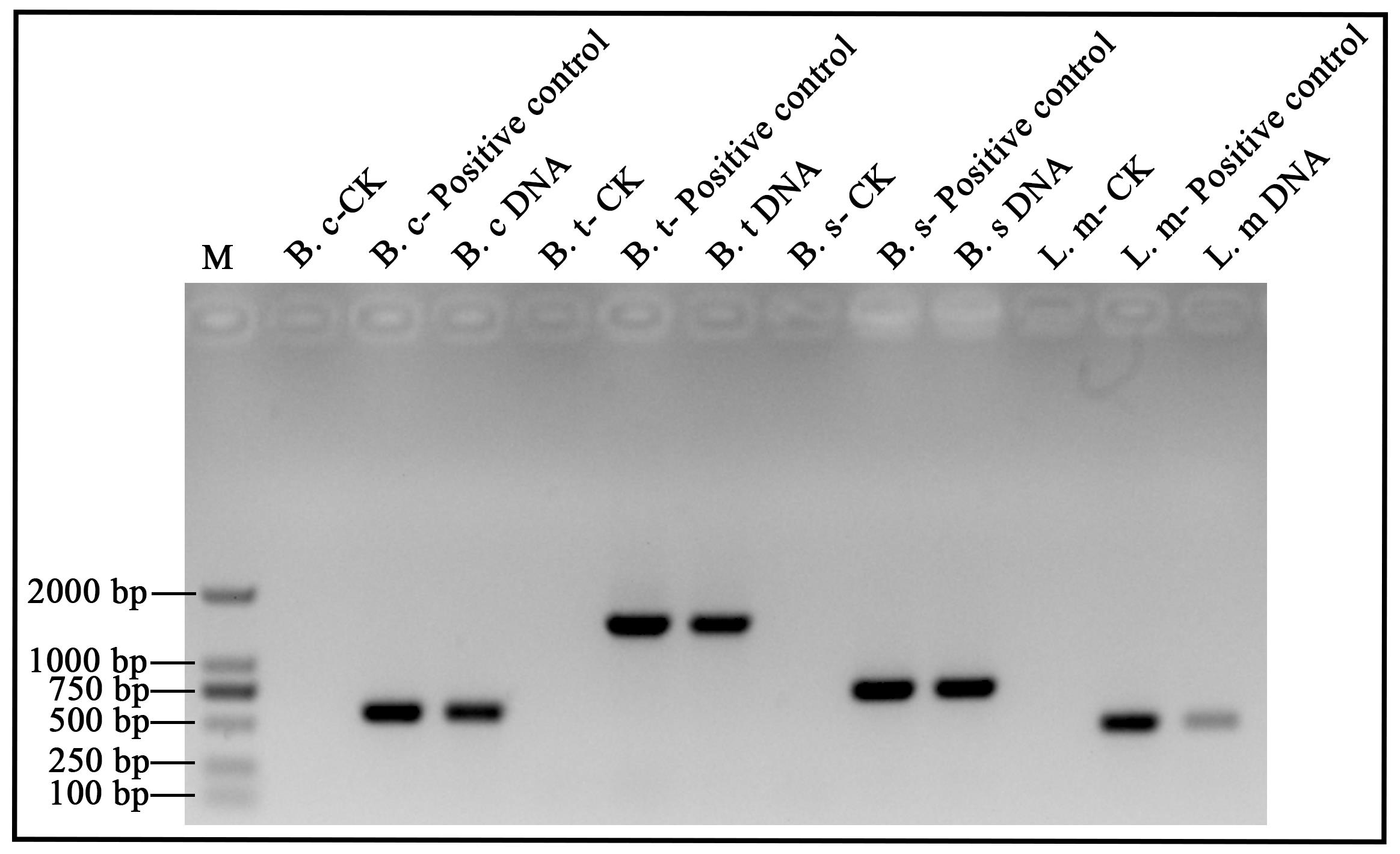
Figure 2. B. cereus, B. thuringiensis, B. subtilis, and L. monocytogenes DNA analyzed by electrophoresis. Lane M: 100–2,000 bp DNA ladder (Marker D). B. c: B. cereus; B. t: B. thuringiensis; B. s: B. subtilis; L. m: L. monocytogenes. DNA: DNA extracts by ultrasonic method for 5 min. Positive control: DNA extracted with a commercial kit (1 μL DNA template). CK: Check control (no template).Table 2. The quality of DNA sample by spectrophotometry analysis
Microorganism Nucleic Acid (ng/μL) A260/A280 A260/A230 B. cereus ATCC 14579 20.343 1.517 0.641 B. thuringiensis ATCC 10792 19.43 1.5 0.605 B. subtilis ATCC 6633 21.935 1.303 0.634 L. monocytogenes ATCC 19115 6.321 1.808 1.018 Table 3. Primer sequences used in B. cereus, B. thuringiensis, L. monocytogenes, and B. subtilis, respectively
Microorganism Target gene Primer sequence (5′ to 3′) B. cereus ATCC 14579 gmk F-TTAAGTGAGGAAGGGTAGG
R-AATGTTCACCAACCACAA
B. thuringiensis ATCC 10792 pycA F-GTGAAAGCAAGAACACAAGC
R-ATAGTTTTTGTATCCAACTGCG
B. subtilis ATCC 6633 SigB F-ATGACACAACCATCAAAAACTACGA
R-TTACATTAACTCCATCGAGGGATCT
L. monocytogenes ATCC 19115 prfA F-GTCAAAACATACGCTCTTATC
R-ACATAATCAGTCCAAAGTAGATGC
Acknowledgments
This protocol was adapted from previous work (Chen et al., 2022; Li et al., 2022). This research was supported by Guangdong Major Project of Basic and Applied Basic Research (2020B0301030005), Guangdong Provincial Key Laboratory (2020B121201009), and Guangdong Province Academy of Sciences Special Project for Capacity Building of Innovation Driven Development (2020GDASYL-20200301002).
Competing interests
There are no conflicts of interest or competing interests.
References
Azevedo, F., Pereira, H. and Johansson, B. (2017). Colony PCR. Methods Mol Biol 1620: 129-139.
- Bello, G. L., Morais, F. C. L., Wolf, J. M., Gehlen, M., Soares, T. D. S., Halon, M. L., Barcellos, R. B. and Rossetti, M. L. R. (2020). Improvement of Mycobacterium tuberculosis detection in sputum using DNA extracted by sonication. Braz J Infect Dis 24(5): 398-404.
- Chen, M., Wei, X., Zhang, J., Zhou, H., Chen, N., Wang, J., Feng, Y., Yu, S., Zhang, J., Wu, S., Ye, Q., Pang, R., Ding, Y. and Wu, Q. (2022). Differentiation of Bacillus cereus and Bacillus thuringiensis Using Genome-Guided MALDI-TOF MS Based on Variations in Ribosomal Proteins. Microorganisms 10(5): 918.
- Han, Z., Sun, J., Lv, A., Sung, Y., Sun, X., Shi, H., Hu, X., Wang, A. and Xing, K. (2018). A modified method for genomic DNA extraction from the fish intestinal microflora. AMB Express 8(1): 52.
- Li, Y., Chen, N., Wu, Q., Liang, X., Yuan, X., Zhu, Z., Zheng, Y., Yu, S., Chen, M., Zhang, J., Wang, J. and Ding, Y. (2022). A Flagella Hook Coding Gene flgE Positively Affects Biofilm Formation and Cereulide Production in Emetic Bacillus cereus. Front Microbiol 13: 897836.
- Mertens, K., Freund, L., Schmoock, G., Hänsel, C., Melzer, F. and Elschner, M. C. (2014). Comparative evaluation of eleven commercial DNA extraction kits for real-time PCR detection of Bacillus anthracis spores in spiked dairy samples. Int J Food Microbiol 170: 29-37.
- Packeiser, H., Lim, C., Balagurunathan, B., Wu, J. and Zhao, H. (2013). An extremely simple and effective colony PCR procedure for bacteria, yeasts, and microalgae. Appl Biochem Biotechnol 169(2): 695-700.
- Taylor, M. T., Belgrader, P., Furman, B. J., Pourahmadi, F., Kovacs, G. T. and Northrup, M. A. (2001). Lysing bacterial spores by sonication through a flexible interface in a microfluidic system. Anal Chem, 73(3), 492–496.
- Yang, Z. H., Xiao, Y., Zeng, G. M., Liu, Y. G. and Deng, J. H. (2006). [DNA extraction methods of compost for molecular ecology analysis]. Huan Jing Ke Xue 27(8): 1613-1617.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Chen, N. and Yuan, X. (2023). A Quick DNA Extraction Method for High Throughput Screening in Gram-positive Bacteria. Bio-protocol 13(8): e4653. DOI: 10.21769/BioProtoc.4653.
Category
Microbiology > Microbial genetics > Transformation
Microbiology > Microbial genetics > DNA > DNA detection and isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








