- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Visualization of Lipid Droplets in the Alveolar Macrophage Cell Line MH-S with Live-cell Imaging by 3D Holotomographic Microscopy (Nanolive)
Published: Vol 13, Iss 5, Mar 5, 2023 DOI: 10.21769/BioProtoc.4629 Views: 1700
Reviewed by: David PaulVasiliki KoliarakiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Rapid and Uniform NHS-Ester-Based Membrane Protein Labeling of Live Mammalian Cells
Alyssa Burgess [...] Ying S. Hu
Oct 5, 2025 2001 Views

Lipid-Mediated Sequential Recruitment of Proteins Via Dual SLIPT and Dual SLIPTNVOC in Live Cells
Kristina V. Bayer and Richard Wombacher
Nov 5, 2025 1484 Views
Abstract
Lipid droplets (LD), triglycerides and sterol esters among them, are well known for their capacity as lipid storage organelles. Recently, they have emerged as critical cytoplasmic structures involved in numerous biological functions. LD storage is generated de novo by the cell and provides an energy reserve, lipid precursors, and cell protection. Moreover, LD accumulation can be observed in some pathologies as obesity, atherosclerosis, or lung diseases. Fluorescence imaging techniques are the most widely used techniques to visualize cellular compartments in live cells, including LD. Nevertheless, presence of fluorophores can damage subcellular components and induce cytotoxicity, or even alter the dynamics of the organelles. As an alternative to fluorescence microscopy, label-free techniques such as stimulated Raman scattering and coherent anti-stokes Raman scattering microscopy offer a solution to avoid the undesirable effects caused by dyes and fluorescent proteins, but are expensive and complex. Here, we describe a label-free method using live-cell imaging by 3D holotomographic microscopy (Nanolive) to visualize LD accumulation in the MH-S alveolar macrophage cell line after treatment with oleic acid, a monounsaturated fatty acid that promotes lipid accumulation.
Keywords: Lipid dropletBackground
Lipid droplets (LD) are dynamic cytoplasmic organelles that serve as intracellular energy storage, mainly in the form of triglycerides and cholesterol esters (van Dierendonck et al., 2022). In addition to their role in lipid storage and transport, LD are increasingly recognized to be involved in the regulation of other cellular processes, including inflammation, cell activation, and metabolism (Xu et al., 2018; Agudelo et al., 2020). Accumulation of LD is also related to several pathologies as obesity, diabetes, atherosclerosis, or lung diseases (Xu et al., 2018; Agudelo et al., 2020). Alveolar macrophages (AM) are the first line of defense against respiratory pathogens and play key roles in lung lipid metabolism (Agudelo et al., 2020). Excessive amounts of intracellular lipids in AM have been described in lung pathologies such as chronic obstructive pulmonary disease, acute lung injury, idiopathic pulmonary fibrosis, or pulmonary alveolar proteinosis, among others (Agudelo et al., 2020). Nevertheless, the mechanisms responsible for LD accumulation are not fully elucidated. Fluorescence-based live-cell imaging is the most widely used technique for LD studies, even though the phototoxicity of fluorescent dyes may disturb LD dynamics. Here, we present a protocol for live imaging of the LD accumulation without the addition of fluorescent labels, using 3D holotomographic microscopy (Nanolive). This label-free microscopy method reports the changes of the refractive indices (RIs) in three dimensions at high spatial and temporal resolution, allowing label-free analysis of organelle biology and kinetics studies with a low level of phototoxicity (Sandoz et al., 2019). We have performed this protocol in the MH-S alveolar macrophage cell line, grown in the presence of oleic acid to induce LD formation. Using this live-cell approach, foam cell formation can be analyzed under a physiological context, excluding the artefacts induced by the addition of different chemicals.
Materials and Reagents
µ-dish cell culture imaging dish, 35 mm, high (Ibidi, catalog number: 81156)
1,000, 200, and 20 µL pipette tips (pre-sterile w/ filter, hinged racks) (ExpellPlus, catalog numbers: 5030150C, 5030090C, 5130062C)
15 mL centrifuge tubes (Corning, catalog numbers: 430791)
Corning® Costar® Stripette® serological pipettes, individually paper/plastic wrapped, 5 and 10 mL (Corning, catalog numbers: CLS4487, 4488)
Falcon® 100 mm TC-treated cell culture dish (Corning, catalog number: 353003)
RPMI 1640 media (Lonza, catalog number: BE 12-115F)
Fetal bovine serum (FBS) (GibcoTM, catalog number: 10270106)
Penicillin-streptomycin mixture (Lonza, catalog number: DE17-603E)
Dulbecco's phosphate buffered saline (PBS) (10×), 95 mM (PO4) without calcium or magnesium (Lonza, catalog number: BE17-515Q)
Bovine serum albumin (BSA)-oleate monounsaturated fatty acid complex (5 mM) (Cayman Chemical, catalog number: 29557)
BSA control for BSA-fatty acid complexes (5 mM) (Cayman Chemical, catalog number: 29556)
Trypsin/EDTA solution (Lonza, catalog number: CC-5012)
Complete RPMI growth medium (see Recipes)
Starving medium (see Recipes)
Biological materials
Cell line: MH-S (ATCC number: CRL-2019TM)
Equipment
Rainin Pipet-Lite XLS (Mettler Toledo, models: SL1000 and SL200)
Forma Direct heat CO2 incubator 184 L digital model (Thermo Scientific, model: 311; TC 230)
Biosafety cabinet (Telstar, model: Bio II Advance Plus IV)
Centrifuge Sorvall ST 16R (Thermo Scientific, model: 75004380)
3D Cell Explorer microscope (Nanolive, Ecublens, Switzerland)
CellDropTM FL automated cell counter (DeNovix)
Laboratory water bath (Memmert, model: WNB 14)
Software
Steve software v1.6.3496 (Nanolive, Ecublens, Switzerland)
ImageJ (FIJI) software (https://imagej.net/software/fiji/downloads)
Procedure
Cell culture
Grow MH-S cells inside of a humidified incubator with 5% CO2 at 37 °C in complete RPMI growth medium (see Recipes) to 90% confluency in a 100 mm dish.
Remove the culture media and wash the cells with 10 mL of PBS 1×.
After removing the PBS, add 2 mL of trypsin/EDTA solution to detach cells, incubate in a humidified incubator with 5% CO2 for 5 min at 37 °C, and add 8 mL of complete RPMI growth medium to stop the reaction.
Note: Temper the growth medium and trypsin solution at 37 °C in a water bath.
Collect the cells and centrifuge at 423 × g for 5 min. Remove supernatant and resuspend the pellet in 5 mL of complete RPMI growth medium.
Calculate the cell concentration using an automated cell counter.
Seed MH-S cells in 35 mm μ-dishes (3 × 105 cells per dish) in 1.5 mL of complete RPMI growth medium.
Note: Remember to seed additional dishes for control condition (see step B3).
Incubate cells at 37 °C in a humidified incubator with 5% CO2 overnight.
Temper the starving medium (see Recipes) at 37 °C in a water bath.
Change the cells to tempered starving medium and incubate under the same culture conditions for approximately 3 h.
BSA-oleate treatment
Defrost an aliquot of BSA-oleate monounsaturated fatty acid complex (5 mM) in the water bath for 2 min.
Prepare the BSA-oleate working solution at 100 µM in starving medium.
Note: For 1.5 mL of medium, you need 60 µL of BSA-oleate monounsaturated fatty acid complex.
Change the starving medium to starving medium + BSA-oleate (100 µM) or starving medium + BSA for the control.
Prewarm the stage of the 3D Cell Explorer microscope to 37 °C at least 30 min before starting the acquisition.
Place the plate in the 3D Cell Explorer microscope stage where the incubation conditions (37 °C and 5% CO2) will be maintained during the rest of the experiment.
Note: Other holotomographic microscopes can also be used.
Image acquisition
This protocol is customized for a 3D Cell Explorer microscope equipped with a 60× magnification dry objective (λ = 520 nm, sample exposure 0.2 mW/mm2) and a depth of field of 30 μm.
Proceed to capture images every minute for 18 hours, using the Steve software that controls the microscope.
Each image provides a reconstruction of 30 μm in the z-axis, which corresponds to 100 images. Most of the images from the 100-image stacks are in fact out of focus, because samples are only visualized in a small range in the z-axis (around 5–10 stacks). From the whole z-stack, select at each time point the z-image in which the focus and resolution of the cells is best observed. LD are observed as intracellular spheres that have a high RI.
Export the image using Steve software to transform RI volumes into .tiff format. Pictures are exported by default as 32-bits images.
Process the exported image (.tiff format) using the FIJI software for performance purposes. Transform images to 8-bits and adjust gamma, brightness, and contrast identically for compared image sets using Fiji software.
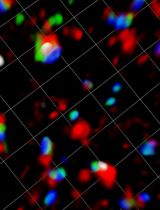
An example of time-lapse refractive images of LD in alveolar macrophages (MH-S cells) is shown in Figure 1.

Figure 1. Time-lapse refractive images of lipid droplets (LD) in alveolar macrophages (MH-S cells). Images show LD accumulation in MH-S cells treated with oleic acid (green) compared to control condition (blue). Close-up images shown at the right column correspond to the position indicated in the small squares.
Recipes
Complete RPMI growth medium
RPMI 1640 media supplemented with 10% FBS and 1% penicillin-streptomycin.
Starving medium
RPMI 1640 media supplemented with 2% FBS and 1% penicillin-streptomycin.
Acknowledgments
We are grateful to Instituto de Salud Carlos III for financial support to S.H.(PI20CIII/00018). A. Pérez-Montero is supported by Comunidad de Madrid (PEJ-2020-AI/BMD-17651).
Competing interests
The authors declare no competing interests.
References
- Agudelo, C. W., Samaha, G. and Garcia-Arcos, I. (2020). Alveolar lipids in pulmonary disease. A review. Lipids in health and disease 19(1): 122-122.
- van Dierendonck, X., Vrieling, F., Smeehuijzen, L., Deng, L., Boogaard, J. P., Croes, C. A., Temmerman, L., Wetzels, S., Biessen, E., Kersten, S., et al. (2022). Triglyceride breakdown from lipid droplets regulates the inflammatory response in macrophages. Proc Natl Acad Sci U S A 119(12): e2114739119.
- Xu, S., Zhang, X. and Liu, P. (2018). Lipid droplet proteins and metabolic diseases. Biochim Biophys Acta Mol Basis Dis 1864(5, Part B): 1968-1983.
- Sandoz, P. A., Tremblay, C., van der Goot, F. G. and Frechin, M. (2019). Image-based analysis of living mammalian cells using label-free 3D refractive index maps reveals new organelle dynamics and dry mass flux. PLoS Biol 17(12): e3000553.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Pérez-Montero, A., Zaragoza, O., Luque, A., Hortelano, S. and Acebo, P. (2023). Visualization of Lipid Droplets in the Alveolar Macrophage Cell Line MH-S with Live-cell Imaging by 3D Holotomographic Microscopy (Nanolive). Bio-protocol 13(5): e4629. DOI: 10.21769/BioProtoc.4629.
Category
Biophysics > Microscopy
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link