- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Assay for Phytaspase-mediated Peptide Precursor Cleavage Using Synthetic Oligopeptide Substrates
Published: Vol 13, Iss 3, Feb 5, 2023 DOI: 10.21769/BioProtoc.4608 Views: 1794
Reviewed by: Giusy TornilloBilly Tasker-BrownSimab Kanwal

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Assessing Metabolite Interactions With Chloroplastic Proteins via the PISA Assay
Anna Karlsson [...] Elton P. Hudson
May 5, 2025 1991 Views
Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1757 Views

Quantitative Analysis of the Arabidopsis Leaf Secretory Proteome via TMT-Based Mass Spectrometry
Sakharam Waghmare [...] Rucha Karnik
Nov 20, 2025 2014 Views
Abstract
Proteases control plant growth and development by limited proteolysis of regulatory proteins at highly specific sites. This includes the processing of peptide hormone precursors to release the bioactive peptides as signaling molecules. The proteases involved in this process have long remained elusive. Confirmation of a candidate protease as a peptide precursor–processing enzyme requires the demonstration of protease-mediated precursor cleavage in vitro. In vitro cleavage assays rely on the availability of suitable substrates and the candidate protease with high purity. Here, we provide a protocol for the expression, purification, and characterization of tomato (Solanum lycopersicum) phytaspases as candidate proteases for the processing of the phytosulfokine precursor. We also show how synthetic oligopeptide substrates can be used to demonstrate site-specific precursor cleavage.
Graphical abstract
Background
Phtytaspases are a subgroup of plant subtilases that are characterized by their specificity for aspartic acid immediately upstream of the cleavage site (in position P1) of their oligopeptide and protein substrates (Chichkova et al., 2014; Schaller et al., 2018). Phytaspases were originally characterized in tobacco and rice (Chichkova et al., 2010) and more recently in Arabidopsis and tomato (Beloshistov et al., 2018; Chichkova et al., 2018; Reichardt et al., 2018 and 2020). In addition to the canonical Asp residue at the scissile bond, several amino acids upstream of the critical Asp were found to contribute to substrate recognition, resulting in high selectivity of individual phytaspases for limited proteolysis at specific sites of their protein targets (Chichkova et al., 2018; Reichardt et al., 2018).
Phytaspases in particular, and plant subtilases in general, are synthesized as pre-pro-proteins that are directed to the secretory pathway for proteolytic maturation, glycosylation, and secretion (Schaller et al., 2018). While there is a single report on the successful expression of phytaspase in E. coli (Narayanan et al., 2017), the complex maturation pattern of subtilases rather calls for an eukaryotic host for recombinant protein expression (Meyer et al., 2016). The baculovirus–insect cell system and stably transformed plant cell lines have been used to produce post-translationally modified and fully active plant subtilases (Janzik et al., 2000; Cedzich et al., 2009; Ottmann et al., 2009). Here, we used Nicotiana benthamiana plants for the expression of C-terminally His-tagged phytaspases by agroinfiltration. This transient expression system may not yield as much recombinant protein as insect and plant cell culture systems but is much more rapid and allows for simple extraction of the secreted subtilases from extracellular (apoplastic) wash fluids. Subsequent purification by metal-chelate affinity chromatography followed by gel filtration results in near homogeneity of the recombinant enzymes for in vitro cleavage assays.
Cleavage assays include synthetic oligopeptides that comprise several precursor-derived amino acids upstream and/or downstream of the cleavage sites as protease substrates. In this experiment, we used a decamer peptide consisting of five amino acids of the precursor followed by the di-sulfated PSK pentapeptide as a substrate for tomato phytaspases. To demonstrate specificity of cleavage, we included a second peptide, in which Ala substituted the critical Asp residue at the cleavage site. Cleavage products were then identified and quantified by mass spectrometry (Reichardt et al., 2020).
We provide a protocol for (A) the transient expression of His-tagged phytaspases by agroinfiltration of N. benthamiana plants, (B) the purification of the recombinant proteins from apoplastic leaf extracts, (C) the cleavage assay using synthetic peptide substrates, and (D) sample preparation for mass spectrometry (MS). Not included is a protocol for the cloning of the protease of interest. In the experiment described here, we used expression constructs for phytaspases from tomato that were generated by conventional cloning techniques in the binary vector pART27 and transformed into Agrobacterium tumefaciens strain C58C1 as described (Reichardt et al., 2018). A protocol for the mass spectrometric analysis of cleavage products is also not included, as this part of the analysis is usually either performed by a central facility of the respective institution or provided as a commercial service. For SDS-PAGE analysis, we followed standard procedures (Stintzi et al., 2022). The protocols we provide here can easily be adapted to other secreted proteases that tolerate the addition of a C-terminal His tag. Peptide sequences will have to be chosen according to the specific substrate requirements of the protease of interest.
Materials and Reagents
50 mL culture tubes (Corning/Falcon, catalog number: 352070)
15 mL culture tubes (Corning/Falcon, catalog number: 352196)
1 mL PE/PP syringes (avantor/VWR, catalog number: 613-2001)
100 mL PE/PP syringes (Th.Geyer/Becton Dickinson, catalog number: 6287774)
Oak Ridge High-Speed PPCO centrifuge tubes (Thermo Fisher Scientific/Nalgene, catalog number 3139-0030)
250 or 500 mL centrifuge bottles (Thermo Fisher Scientific/Nalgene, catalog number: 3141-0250 or 3141-0500)
Scalpel or razor blades
17 or 18 gauge blunt-tipped syringe needle (e.g., B. Braun Sterican®, 18G / 1.2 × 40 mm, catalog number: 4038088)
300 or 500 mL Pyrex beakers
Vivaspin 20 centrifugal concentrator (30 k molecular weight cut-off) (Sartorius Stedim, catalog number: VS2021)
A. tumefaciens C58C1 (RifR, TetR) (community resource; NCBI:txid176299)
Expression construct for the protease of interest under control of the CaMV 35S promoter in a binary vector for plant transformation. Here, we used expression constructs for His-tagged tomato phytaspases in pART27 (SpecR) transformed into A. tumefaciens, strain C58C1 (Reichardt et al., 2018). Agrobacteria transformed with the empty expression vector (here pART27) were used as control
A. tumefaciens C58C1 with p19 silencing suppressor in pBin61 (KanR, RifR, TetR) (Voinnet et al., 2003)
Nicotiana benthamiana seeds (Agroscience GmbH, Neustadt, Germany)
N. benthamiana plants, grown on seeding substrate at 25°C and 16:8 h day/night cycle. Plants older than three weeks are watered with 1.48 g/L N (20%), P (20%), and K (20%) universal fertilizer including micronutrients.
Note: Expression levels are much reduced in flowering plants. Therefore, use the plants before they start to bolt, usually when they are between four and five weeks old
Spectinomycin (Duchefa, catalog number: S0188); 100 mg/mL stock solution in H2O, store at -20°C
Rifampicin (Duchefa, catalog number: R0146); 100 mg/mL stock solution in DMSO, store at -20°C
Tetracycline (Duchefa, catalog number: T0150); 25 mg/mL stock solution in 70 % (v/v) ethanol, store at -20°C
Kanamycin sulphate (Duchefa, catalog number: K0126); 50 mg/mL stock solution in H2O, store at -20°C
Tryptone (Duchefa, catalog number: T1332)
Nickel (Ni)-NTA agarose (Qiagen, catalog number: 30210), store at 4°C
Bio-Rad Protein Assay kit II, with bovine serum albumin as the standard protein (Bio-Rad, catalog number: 5000002)
Coomassie protein stain (e.g., InstantBlue, abcam, catalog number: ISB1L)
Custom-synthesized synthetic substrate peptides at >90% purity, EAHLD[sY]I[sY]TQM and EAHLA[sY]I[sY]TQM (sY = sulfotyrosine), (PepMic, Suzhou, China). The lyophilized peptides are stored at -20°C. Working solutions can be stored at 4°C for one month, or at -20°C. Avoid repeated freeze/thaw cycles
MgCl2·6H2O (Carl Roth, catalog number: 3532.1)
NaCl (Carl Roth, catalog number: 9265.2)
KCl (Carl Roth, catalog number: 6781.1)
NaH2PO4·2H2O (Carl Roth, catalog number: T879.2)
Na2HPO4·2H2O (Carl Roth, catalog number: T877.1)
NaOH (Carl Roth, catalog number: P031.3)
Acetosyringone (Carl Roth, catalog number: 6003.2), store at -20°C
Imidazole (Carl Roth, catalog number: 3899.4)
2-(N-Morpholino)-ethanesulfonic acid (MES·H2O) (Carl Roth, catalog number: 6066.2)
Yeast extract (Carl Roth, catalog number: 2904.3)
Glacial acetic acid (Carl Roth, catalog number: 7332.1)
Glycerol (Carl Roth, catalog number: 4043.1)
Acetonitrile (Carl Roth, catalog number: 7330.2)
Trifluoroacetic acid (Carl Roth, catalog number: P088.1)
H2O, HPLC grade (Carl Roth, catalog number: A511.2)
LB medium (lysogeny broth) (see Recipes)
Infiltration buffer (see Recipes)
Extraction buffer, reaction buffer (see Recipes)
Binding buffer (see Recipes)
Elution buffer (see Recipes)
Gel filtration buffer (see Recipes)
Solvent A (see Recipes)
Solvent B (see Recipes)
Solvent C (see Recipes)
Equipment
Microcentrifuge (Eppendorf, model: 5418 R)
Tabletop centrifuge with swing-out rotor for 15/50 mL tubes (Eppendorf, model: 5810 R)
RC-3B refrigerated centrifuge (Sorvall Instruments) for 250/500 mL bottles
RC6+ refrigerated centrifuge (Sorvall Instruments) for 30 mL tubes
UV/Vis spectrophotometer (e.g., Eppendorf Biophotometer, catalog number: 634-0839)
Desiccator (5 L) with vacuum pump
Rotating wheel (e.g., Steinberg Systems, model: SBS-LBM-200)
Microbiological incubators for plate cultures at 28 and 37°C
Enrich SEC 650 10 × 300 gel filtration column (Bio-Rad, catalog number: 7801650)
Fast protein liquid chromatography system. We used the NGC Quest system (Bio-Rad, catalog number: 788-0003)
Note: Other chromatography systems, e.g., ÄKTA Pure (Cytiva, catalog number: 29018226) can be used as well.
Vacuum concentrator, e.g., Savant SpeedVac (Thermo Fisher Scientific, catalog number: SPD1030A)
Standard SDS-PAGE equipment (e.g., Mini-PROTEAN Tetra Cell and casting module from Bio-Rad, catalog numbers: 1658000EDU and 1658015EDU)
Electrophoresis power supply (e.g., PowerPac Basic from Bio-Rad, catalog number: 1645050)
PTFE (polytetrafluorethylene; TeflonTM) membranes with embedded C18 beads, e.g., Supelco EmporeTM solid phase extraction discs (Millipore Sigma/Supelco, catalog number: 66887U)
Procedure
Transient expression of phytaspases in N. benthamiana leaves
Pick single colonies of A. tumefaciens C58C1 containing the phytaspase expression vector or the empty-vector control and resuspend them in 500 µL of LB medium.
Streak the resuspended colonies evenly on LB plates with appropriate antibiotics (rifampicin, 100 µg/mL; tetracycline, 25 µg/mL; spectinomycin, 100 µg/mL).
Note: The goal is to get a bacterial lawn.
Similarly, streak a single colony of agrobacteria carrying the P19 expression construct resuspended in 500 µL of LB on LB plates containing kanamycin (50 µg/mL) instead of spectinomycin.
Incubate for 48 h at 28°C.
Use 6 mL of infiltration buffer (see Recipes) to wash off the bacteria from the respective plates and transfer the suspensions into 15 mL culture tubes.
Collect cells by centrifugation for 10 min at 1,000 × g; discard supernatant.
Resuspend cells in 6 mL of infiltration buffer each.
Determine optical density (OD) at 600 nm.
Mix the two suspensions in a 50 mL culture tube and dilute with infiltration buffer, to obtain 50 mL of infiltration solution with a final OD600 of 0.7 for the phytaspase expression construct and 1:0 for the P19 silencing suppressor.
Do the same for the empty-vector control.
Use a 1 mL plastic syringe without the needle to inject the suspensions into the abaxial side of leaves of N. benthamiana plants (Figure 1A, Video 1).
Note: Support the leaf on the opposite site with your index finger. Use gentle pressure and be careful not to injure the leaves. Infiltrated areas are water-soaked; they become much darker than the remainder of the leaf. Try to infiltrate as much of the leaf area using as few infiltration sites as possible.
Video 1. Agroinfiltration of N. benthamiana plantsContinue to grow the plants at 25°C and 16:8 h day/night cycle.
Harvest the leaves at five days after infiltration.
Note: The optimum time for transient protein expression in N. benthamiana depends on the protein of interest and needs to be determined experimentally. If degradation of the expressed protease or tissue necrosis as a result of protease overexpression are observed, earlier time points may have to be chosen for maximum yield.
Use a razor blade or scalpel to remove the central vein.
Place the remaining leaf material, upper side down, into a beaker (one for the expression construct and one for the empty-vector control) containing approximately 100 mL of extraction buffer (see Recipes) on ice (Figure 1B).
Place the beakers into a desiccator and vacuum infiltrate the extraction buffer at 75 mbar for 2 min; then, slowly release the vacuum.
Blot the leaves dry (Figure 1C). Stack them on top of each other, roll them up (Figure 1D), and place them into the barrel of a 100 mL plastic syringe (Figure 1E).

Figure 1. Transient protein expression in N. benthamiana leaves. A. Infiltration of agrobacterial suspension into the abaxial side of N. benthamiana leaves. B. Harvesting of leaves into extraction buffer. C. Leaves on blotting paper after vacuum infiltration. D, E. Leaves are piled one on top of the other and rolled up (D), in order to place them into the barrel of a 100 mL plastic syringe (E). (F) Diagram of a syringe barrel placed into a centrifuge bottle.Place the syringe barrel into a 250/500 mL centrifuge bottle (Figure 1E, F) and spin at 1,500 × g for 7 min at 4°C, to collect the intercellular fluid (apoplastic wash).
Transfer the wash fluid to Oak Ridge centrifuge tubes and clear by centrifugation at 20,000 × g at 4°C.
Transfer the supernatant to a new tube
Add imidazole to a final concentration of 4 mM.
Purification of phytaspases
His-tagged phytaspases are purified from apoplastic washes by metal-chelate affinity chromatography on Ni-NTA agarose in a batch procedure, followed (optionally) by size exclusion chromatography on an Enrich SEC 650 10 × 300 gel filtration column. As a control, apoplastic washes from empty-vector-infiltrated plants are subjected to the same purification procedure.
Pipette 500 µL of Ni-NTA agarose (50% slurry of agarose beads in storage buffer) into a 15 mL culture tube and spin for 2 min at 500 × g to sediment the matrix. Prepare two tubes, one for the recombinant protease and one for the mock purification from empty-vector-infiltrated plants.
To wash the matrix, use a pipette to remove the buffer. Note: Be careful not to aspirate the settled matrix. Add 10 mL of binding buffer (see Recipes), mix gently and spin down, and remove the buffer as before. Repeat two times. Resuspend in 1 mL of binding buffer after the final wash.
Add the apoplastic wash from step A19 to the matrix and incubate for 1 h at 4°C on a rotating wheel.
Collect the matrix by centrifugation as above and wash three times with 12 mL of binding buffer, as described in step B2.
Carefully remove the buffer after the last washing step.
Add 600 µL of elution buffer (see Recipes), sediment by centrifugation, and recover the supernatant. Repeat twice.
Combine the three 600 µL eluates and reduce the volume to approximately 100 µL by ultrafiltration using a Vivaspin centrifugal concentrator (30 k molecular weight cut-off) according to the manufacturer’s instructions.
For buffer exchange, add 500 µL of elution buffer without imidazole. Reduce the volume to 100 µL as before (step B7). Repeat three times.
Assess purity by SDS-PAGE and Coomassie-Brilliant Blue staining following standards protocols (Stintzi et al., 2022) (Figure 2A).
Note: Many proteases that are expressed at high levels are sufficiently pure for further characterization at this stage (see for example P69A in Figure 2A). However, for applications that require the highest purity (e.g., the PICS assay for analysis of substrate specificity, as reported in Reichardt et al., 2018), we recommend further purification by gel filtration as detailed in the following steps B10–B14.
Apply the sample to an Enrich SEC 650 10 × 300 gel filtration column (or similar), equilibrated in 50 mM sodium phosphate buffer pH 7.0, 300 mM NaCl, at 0.5 mL/min on an NGC Quest (Bio-Rad) chromatography system.
Monitor UV absorbance at 280 nm; collect the column eluate in 200 µL fractions. Results are shown for tomato phytaspase 2 (Phyt2) in Figure 2B.
For the highest purity, use only the fraction at the peak maximum in further experiments. Alternatively, pool all peak fractions, concentrate by ultrafiltration as above (step B7), add glycerol to 50% final concentration, and store at -20°C.
Determine protein concentration using a commercial Bradford assay system (e.g., Bio-Rad Protein Assay kit II) following the manufacturer’s instructions.
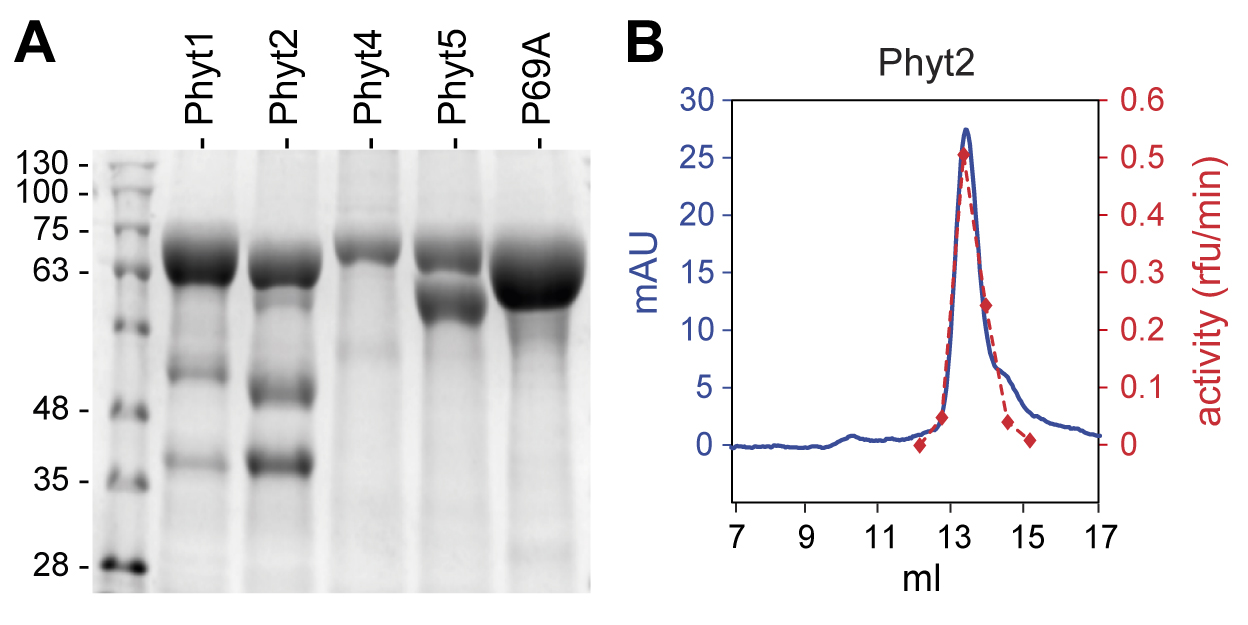
Figure 2. Purification of tomato phytaspases from apoplastic extracts of agroinfiltrated N. benthamiana plants. A. SDS-PAGE analysis of phytaspases purified from apoplastic extracts by metal-chelate affinity chromatography on Ni-NTA agarose. Approximately 2 (Phyt4), 5 (Phyt5), or 6 µg (Phyt1, Phyt2, P69A) of protein were loaded onto the gel. A Coomassie-stained 10% gel is shown; the molecular mass of marker proteins is indicated in kDa. B. Further purification of Phyt2 by gel permeation chromatography. The elution volume is shown in milliliters. Protein elution was monitored at 280 nm and is shown as m(illi) A(bsorbance) U(nits) in blue; 200 µL fractions were collected and assayed for Phyt2 activity using a fluorogenic peptide substrate. Activity is shown in arbitrary units in red, as relative fluorescence increase per minute. Modified from Reichardt et al. (2018), Figures 3a and 4a (Reichardt et al., 2018).
In vitro cleavage assay for phytaspase specificity
Activity and cleavage specificity of the purified protease of interest is tested with synthetic oligopeptides as substrates. Lyophilized custom-synthesized peptides can be obtained at >90% purity from commercial suppliers. The amino acid sequence has to be chosen to match the substrate requirements of the protease of interest. To analyze the cleavage specificity of tomato phytaspases, we used a decamer peptide comprising the five residues of PSK ([sY]I[sY]TQM; sY = sulfotyrosine) with five additional precursor-derived amino acids at the N-terminus (EAHLD[sY]I[sY]TQM) and a second peptide in which the critical Asp residue at the cleavage site was replaced by Ala (EAHLA[sY]I[sY]TQM).
Resuspend the lyophilized peptide in ddH2O; determine the concentration spectrophotometrically at 260 nm, based on the molar extinction coefficient for two sulfotyrosine residues (ϵ260 = 566 M-1 cm-1).
Set up the in vitro digest in 100 µL of reaction buffer (see Recipes). Prepare two tubes, one containing 140 nM of the purified phytaspase, and the second an equal volume of the mock-purified fraction.
Note: Use high-quality microfuge tubes (e.g., the original Eppendorf tubes) and tips at this and subsequent steps of protocol C. Low quality tubes may leak plasticizer into the sample, which is detrimental to MS/MS analysis.
Start the reaction by adding 5 µM substrate peptide and incubate at 30°C.
Note: The peptide substrates were used in a 3,000-fold molar excess over the protease. However, this cannot be generalized. The quantity of protease in the assay and the required substrate concentration depend on the properties of the protease under study and have to be adjusted accordingly.
Prepare six microfuge tubes, each containing 90 µL of 0.1% trifluoroacetic acid (TFA).
Stop the reaction by taking 10 µL aliquots at 0 min, 10 min, 30 min, 1 h, 5 h, 24 h. Add aliquots to the tubes containing 90 µL of 0.1% TFA.
Sample preparation for mass spectrometry
Prior to mass spectrometry (nanoLC-ESI-MS/MS) analysis, samples need to be desalted and concentrated on C18 ZIP tips or StageTips (Rappsilber et al., 2003).
In order to prepare StageTips, use a hypodermic needle to punch out small disks from PTFE membranes with embedded C18 beads, and place them into 200 µL (yellow) pipet tips. Use two discs per tip. Full details for StageTip preparation are given in Rappsilber et al. (2003).
To condition the StageTips, add 50 µL of solvent A (see Recipes) and spin in a microfuge tube at 2,300 × g for 1 min at 4°C.
Wash twice by adding 100 µL of solvent B (see Recipes) and spin as above.
Apply the sample from step C5 and spin at 800 × g for 1 min at 4°C.
Wash twice by adding 150 µL of solvent B and spin at 2,300 × g for 1 min at 4°C.
Transfer the StageTips to new microfuge tubes and elute twice by adding 20 µL of solvent C (see Recipes); centrifuge at 800 × g for 1 min at 4°C.
Dry samples in a vacuum concentrator and continue with nanoLC-ESI-MS/MS analysis or store at -20°C until further analysis.
Data analysis
Phytaspases are Asp-specific proteases. They cleave their substrate proteins on the carboxy side of aspartic acid residues. The peptide derived from the PSK precursor, EAHLD[sY]I[sY]TQM, that was tested here as a phytaspase substrate is thus expected to be cleaved between aspartate (D) and sulfotyrosine (sY). To confirm Asp-specificity of cleavage, we used a second peptide, in which the critical Asp residue was replaced by Ala. The Ala-substituted peptide, EAHLA[sY]I[sY]TQM, is expected not to be cleaved by phytaspases or to be much less efficiently cleaved as compared to the original peptide. Search the MS/MS data for any cleavage products generated from the two substrate peptides. As search parameters, do not specify a specific enzyme and set mass tolerance at 5 ppm for peptide precursors and 0.02 Da for fragment ions (Note: The mass tolerance settings may vary depending on the resolution and mass accuracy of the mass spectrometer used). The sulfotyrosine side chain is not very stable, resulting in the loss of sulfate, and methionine is frequently oxidized during MS analysis. Therefore, allow for methionine oxidation and tyrosine sulfation as variable modifications during the MS search. Scrutinize the search results for peptides diagnostic for cleavage at the D-sY bond. These are the N-terminal cleavage products EAHLA and EAHLD for the Ala-substituted and the original peptide, respectively, and the C-terminal cleavage product [sY]I[sY]TQM that is the same for both substrate peptides. Be aware that the C-terminal cleavage product contains two sulfotyrosine and one methionine residue, resulting in many possible variants. In our analysis of Phyt2 cleavage specificity, we therefore used the N-terminal peptide for quantification. We quantified the EAHLD and EAHLA cleavage products as the sum of ion intensities for the MS/MS fragment ions of the b- and y-series. Results shown by Reichardt et al. (2020) in Figure 4 and in Supplementary Figure S11 indicated that the PSK-derived peptide is cleaved more efficiently compared to its Ala-substituted variant. EAHLA-derived fragment ions amounted to 20% ± 8% of the corresponding EAHLD-derived ions, which is significantly different from the 1-to-1 ratio expected if both precursor ions were cleaved with equal efficiency.
Recipes
LB medium (lysogeny broth)
Reagent Final concentration Amount Tryptone 10 g/L 10 g Yeast extract 5 g/L 5 g NaCl
Agar (only for solid media)
H2O
5 g/L
15 g/L
n/a
5 g
15 g
up to 1 L
Autoclave for 20 min. Cool down to 60°C and then add antibiotics from 1,000× stock solutions. Use for liquid culture or pour plates if solid media are needed.
Infiltration buffer
Reagent Final concentration Amount MgCl2 (1 M) 10 mM 1 mL MES-NaOH (0.5 M, pH 5.6) 10 mM 2 mL Acetosyringone (100 mM)
H2O
0.15 mM
n/a
1.5 mL
95.5 mL
Total n/a 100 mL Extraction buffer, reaction buffer
Reagent Final concentration Amount NaH2PO4/Na2HPO4 (1 M, pH 6.5) 50 mM 10 mL KCl (1 M)
H2O
200 mM
n/a
40 mL
150 mL
Total n/a 200 mL Binding buffer
Reagent Final concentration Amount NaH2PO4/Na2HPO4 (1 M, pH 7.0) 50 mM 10 mL KCl (1 M)
Imidazole (1 M)
H2O
200 mM
4 mM
n/a
40 mL
0.8 mL
149.2 mL
Total n/a 200 mL Elution buffer
Reagent Final concentration Amount NaH2PO4/Na2HPO4 (1 M, pH 7.0) 50 mM 5 mL KCl (1 M)
Imidazole (1 M)
H2O
200 mM
200 mM
n/a
20 mL
20 mL
55 mL
Total n/a 100 mL Gel filtration buffer
Reagent Final concentration Amount NaH2PO4/Na2HPO4 (1 M, pH 7.0) 50 mM 50 mL NaCl (1 M) 300 mM 300 mL H2O
Total
n/a
n/a
650 mL
1,000 mL
Solvent A
Reagent Final concentration Amount Glacial acetic acid 0.5% (v/v) 0.5 mL Acetonitrile 80% (v/v) 80 mL H2O, HPLC-grade
Total
n/a
n/a
19.5 mL
100 mL
Solvent B
Reagent Final concentration Amount Glacial acetic acid 0.5% (v/v) 0.5 mL H2O, HPLC-grade
Total
n/a
n/a
99.5 mL
100 mL
Solvent C
Reagent Final concentration Amount Glacial acetic acid 0.5% (v/v) 0.5 mL Acetonitrile 50% (v/v) 50 mL H2O, HPLC-grade
Total
n/a
n/a
49.5 mL
100 mL
Acknowledgments
The original research papers in which these procedures were described (Reichardt et al., 2018 and 2020); Research in our laboratory is supported by the German Research Foundation (DFG).
Competing interests
No financial or non-financial competing interests are declared.
References
- Beloshistov, R. E., Dreizler, K., Galiullina, R. A., Tuzhikov, A. I., Serebryakova, M. V., Reichardt, S., Shaw, J., Taliansky, M. E., Pfannstiel, J., Chichkova, N. V., et al. (2018). Phytaspase-mediated precursor processing and maturation of the wound hormone systemin. New Phytol 218(3): 1167-1178.
- Cedzich, A., Huttenlocher, F., Kuhn, B. M., Pfannstiel, J., Gabler, L., Stintzi, A. and Schaller, A. (2009). The protease-associated (PA) domain and C-terminal extension are required for zymogen processing, sorting within the secretory pathway, and activity of tomato subtilase 3 (SlSBT3). J Biol Chem 284(21): 14068-14078.
- Chichkova, N. V., Galiullina, R. A., Beloshistov, R. E., Balakireva, A. V. and Vartapetian, A. B. (2014). Phytaspases: Aspartate-specific proteases involved in plant cell death. Russ J Bioorg Chem 40(6): 606-611.
- Chichkova, N. V., Galiullina, R. A., Mochalova, L. V., Trusova, S. V., Sobri, Z. M., Gallois, P. and Vartapetian, A. B. (2018). Arabidopsis thaliana phytaspase: identification and peculiar properties. Funct Plant Biol 45(2): 171-179.
- Chichkova, N. V., Shaw, J., Galiullina, R. A., Drury, G. E., Tuzhikov, A. I., Kim, S. H., Kalkum, M., Hong, T. B., Gorshkova, E. N., Torrance, L., et al. (2010). Phytaspase, a relocalisable cell death promoting plant protease with caspase specificity. EMBO J 29(6): 1149-1161.
- Janzik, I., Macheroux, P., Amrhein, N. and Schaller, A. (2000). LeSBT1, a subtilase from tomato plants. Overexpression in insect cells, purification and characterization. J Biol Chem 275(7): 5193-5199.
- Meyer, M., Leptihn, S., Welz, M. and Schaller, A. (2016). Functional characterization of propeptides in plant subtilases as intramolecular chaperones and inhibitors of the mature protease. J Biol Chem 291(37): 19449-19461.
- Narayanan, S., Sanpui, P., Sahoo, L. and Ghosh, S. S. (2017). Heterologous expression and functional characterization of phytaspase, a caspase-like plant protease. Int J Biol Macromol 95: 288-293.
- Ottmann, C., Rose, R., Huttenlocher, F., Cedzich, A., Hauske, P., Kaiser, M., Huber, R. and Schaller, A. (2009). Structural basis for Ca2+-independence and activation by homodimerization of tomato subtilase 3. Proc Natl Acad Sci U S A 106(40): 17223-17228.
- Rappsilber, J., Ishihama, Y. and Mann, M. (2003). Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75(3): 663-670.
- Reichardt, S., Piepho, H.-P., Stintzi, A. and Schaller, A. (2020). Peptide signaling for drought-induced tomato flower drop. Science 367(6485): 1482-1485.
- Reichardt, S., Repper, D., Tuzhikov, A. I., Galiullina, R. A., Planas-Marques, M., Chichkova, N. V., Vartapetian, A. B., Stintzi, A. and Schaller, A. (2018). The tomato subtilase family includes several cell death-related proteinases with caspase specificity. Sci Rep 8(1): 10531.
- Schaller, A., Stintzi, A., Rivas, S., Serrano, I., Chichkova, N. V., Vartapetian, A. B., Martínez, D., Guiamét, J. J., Sueldo, D. J., van der Hoorn, R. A. L., et al. (2018). From structure to function – a family portrait of plant subtilases. New Phytol 218(3): 901-915.
- Stintzi, A., Stührwohldt, N., Royek, S., Schaller, A. (2022). Identification of cognate protease/substrate pairs by use of class-specific inhibitors. In: Klemenčič, M., Stael, S., Huesgen, P. F. (Eds.). Plant Proteases and Plant Cell Death. Methods in Molecular Biology, vol 2447. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2079-3_6.
- Voinnet, O., Rivas, S., Mestre, P. and Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33(5): 949-956.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Reichardt, S., Stintzi, A. and Schaller, A. (2023). Assay for Phytaspase-mediated Peptide Precursor Cleavage Using Synthetic Oligopeptide Substrates. Bio-protocol 13(3): e4608. DOI: 10.21769/BioProtoc.4608.
- Reichardt, S., Piepho, H.-P., Stintzi, A. and Schaller, A. (2020). Peptide signaling for drought-induced tomato flower drop. Science 367(6485): 1482-1485.
Category
Plant Science > Plant biochemistry > Protein
Biochemistry > Protein > Synthesis
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










