- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Methods to Detect AUTOphagy-Targeting Chimera (AUTOTAC)-mediated Targeted Protein Degradation in Tauopathies
(*contributed equally to this work) Published: Vol 13, Iss 2, Jan 20, 2023 DOI: 10.21769/BioProtoc.4594 Views: 3015
Reviewed by: Gal HaimovichHyeong-Reh Choi KimNingfei An

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Simplified Paradigm of Late Gestation Transient Prenatal Hypoxia to Investigate Functional and Structural Outcomes from a Developmental Hypoxic Insult
Elyse C. Gadra and Ana G. Cristancho
Oct 5, 2022 2068 Views
Real-Time Autophagic Flux Measurements in Live Cells Using a Novel Fluorescent Marker DAPRed
Arnold Sipos [...] Edward D. Crandall
Mar 5, 2024 2419 Views

Detection of Autophagy in Human Peripheral Blood Mononuclear Cells Using Guava® Autophagy and Flow Cytometry
Melanie Scherer [...] Jörg Bergemann
Sep 20, 2025 1411 Views
Abstract
Targeted protein degradation (TPD) facilitates the selective elimination of unwanted and pathological cellular cargoes via the proteasome or the lysosome, ranging from proteins to organelles and pathogens, both within and outside the cell. Currently, there are several in vitro and in vivo protocols that assess the degradative potency of a given degrader towards a myriad of targets, most notably soluble, monomeric oncoproteins. However, there is a clear deficiency of methodologies to assess the degradative potency of heterobifunctional chimeric degraders, especially those in the autophagy space, against pathological, mutant tau species, such as detergent-insoluble oligomers and high-molecular aggregates. The protocol below describes both in vitro and in vivo biochemical assays to induce tau aggregation, as well as to qualitatively and quantitatively measure the degradative potency of a given degrader towards said aggregates, with specific applications of the AUTOTAC (AUTOphagy-TArgeting Chimera) platform provided as an example. A well-defined set of methodologies to assess TPD-mediated degradation of pathological tau species will help expand the scope of the TPD technology to neurodegeneration and other proteinopathies, in both the lab and the clinic.
Graphical abstract

Overview of assays observing elimination of tauP301L aggregates with AUTOTAC. (A) Description of the biological working mechanism of heterobifunctional chimeric AUTOTAC degraders. (B) Schematic illustration of assays described in this paper.
Background
Targeted protein degradation (TPD) is a global protein editing tool with the potential to modulate the half-life of any proteinaceous cargoes (Chamberlain and Hamann, 2019). In addition to serving as a research tool for protein degradation and function, TPD offers an alternative—and thus attractive—means to eradicate disease-causing agents, especially those entrenched in the previously-considered undruggable proteome (Moon and Lee, 2018; Fisher and Phillips, 2018; Schapira et al., 2019; Ji et al., 2022). The various TPD platforms to date harness either the proteasome or the lysosome to facilitate the targeted elimination of a wide variety of cargoes, both within and outside the cell via distinct mechanisms, exemplified by PROteolysis-Targeting Chimera (PROTAC), Antibody-based proTAC (AbTAC), Lysosome-Targeting Chimera (LYTAC), Autophagy-Targeting Chimera (AUTAC), and AuTophagosome-TEthering Compound (ATTEC), to name just a few (Takalo et al., 2013; Thibaudeau et al., 2018; Takahashi et al., 2019; Li et al., 2019; Banik et al., 2020).
Despite the rapid development of TPD platforms with distinct mechanisms of action, the large majority of focus has been centered around a relatively few number of targets, mostly oncoproteins. Even with the emergence of autophagy/lysosome-based degraders, efficient degradation has been limited to only a handful of alternative targets—especially those responsible for neurodegeneration and proteopathies in general. Consequently, development of novel autophagy-based degraders that can target a wide range of pathological hallmark proteins of neurodegeneration and proteopathies, most notably their high-molecular weight, oligomeric, aggregated, and amyloidogenic species, still remains a pressing issue. A notable obstacle to such development is the relative lack of in vitro and in vivo methodologies to analyze in depth the degradative potency and mechanism of action of such degraders. Such difficulty is exacerbated by the fact that, unlike oncoproteins, the downstream signaling cascades of hallmark proteins of neurodegeneration/proteopathies still remain murky, which accentuates even further the importance of well-defined, precise assays and models to assess the autophagic mechanism of action and potency of an autophagy-based degrader (Peeraer et al., 2015; Lim et al., 2018).
To that end, we have recently reported the development and proof-of-concept characterization of a novel p62-targeting/activating TPD platform called AUTOphagy-TArgeting Chimera (Ji et al., 2022). AUTOTAC degraders can selectively recognize and target a broad range of cellular proteins to autophagic membranes for lysosomal degradation, through the selective interaction and activation of the archetypal autophagy cargo receptor p62/SQSTM1 via its ZZ domain. This interaction induces the conformational activation of inactive p62 into an autophagy-compatible form, which exposes PB1 and LIR domains, and facilitates p62 self-oligomerization in complex with targets and its interaction with LC3 on autophagic membranes (Cha-Molstad et al., 2015; Cha-Molstad et al., 2017; Ji et al., 2017, 2019, 2020; Zhang et al., 2018; Yoo et al., 2018; Heo et al., 2021, 2022). AUTOTAC technology was shown to efficiently degrade both oncoproteins and misfolded, aggregation-prone proteins of neurodegeneration.
In this paper, adopted from our paper describing the proof-of-concept development of the AUTOTAC platform, we describe both in vitro methods and in vivo models to not only induce pathological tau aggregation but also assess the selective degradative potency of a given TPD platform towards said aggregates and fibrils (Ji et al., 2022). To this end, we employed SH-SY5Y-hTauP301L cell lines and murine model that can utilize various fluorescent reporters, including full or split halves of the GFP protein, as well as a tandem mRFP-GFP fusion (Shin et al., 2020). Using these models, we induced the formation of detergent-insoluble, high-molecular weight aggregates of the mutant recombinant tau both in vitro and in vivo. The AUTOTAC TPD platform was shown to successfully degrade these pathological tau species in a lysosome-dependent manner. Overall, the protocol provides a systematic approach to generating pathological tau for the testing of not only AUTOTACs but also other autophagy-based degraders and small-molecule compounds in general, which may be expanded to other hallmark proteins of misfolding and aggregation-prone proteopathies and neurodegeneration.
Materials and Reagents
Part I. In vitro degradation of tauP301L aggregates
5 mL pipette aid (SPL, catalog number: 91010)
10 mL pipette aid (SPL, catalog number: 91005)
1.5 mL e-tube (Axygen, catalog number: MCT-150-C)
100 mm dish (Nest, catalog number: 704002, SPL, catalog number: 30100)
6-well plate (Nest, catalog number: 703003, SPL, catalog number: 30006)
12-well plate (Nest, catalog number: 712003, SPL, catalog number: 30012)
24-well plate (Nest, catalog number: 702002, SPL, catalog number: 30024)
Kim-wipe or paper towel (Yuhan-Kimberly, catalog number: 41112)
Polyvinylidene difluoride membrane, 0.45 μm (Millipore, catalog number: IPVH00010)
X-ray film (AGFA, catalog number: CP-BU)
Microscope slide (Marienfield, catalog number: 1000612)
Cover glass (Marienfield, catalog number: 0117520)
PBS (Biosesang, catalog number: P2007-1)
DMEM (Life Technologies, Gibco®, catalog number: 11995-065)
RPMI (Life Technologies, Gibco®, catalog number: 22400-089)
Opti-mem (Life Technologies, Gibco®, catalog number: 31984-070)
0.05% trypsin (Life Technologies, Gibco®, catalog number: 25300)
Lipofectamine 2000 (Invitrogen, catalog number: 11668019)
Sodium dodecyl sulfate (SDS) (Duchefa Biochemie, catalog number: S1377.1000)
Triple-distilled water
1 M Tris-HCl, pH 6.8 (Biosesang, catalog number: TR2016-050-68)
1.5 M Tris-HCl, pH 8.8 (Biosesang, catalog number: TR4020-050-88)
APS (Ammonium Persulfate) (Bio-Rad, catalog number: 1610700)
TEMED (Bio-Rad, catalog number: 161-0801)
2-Propanol (Sigma-Aldrich, catalog number: L53191334)
5× Laemmli sample buffer (Elpis Biotech, catalog number: EBA-1052)
Skim milk (Georgiachem, catalog number: SM2010)
SuperSignalTM West PICO PLUS (Thermo Fisher Scientific, catalog number: 34578)
PFF (Recombinant Human Tau P301L pre-formed fibrils) (NOVUS, catalog number: NBP2-76794)
Hydroxychloroquine (HCQ) (Sigma, catalog number: A25547)
Okadaic acid (Enzo, catalog number: ALX-350-003-C100)
HEPES (Duchefa Biochemie, catalog number: H1504.011)
PierceTM protein transfection reagent (PJ) (Thermo Fisher Scientific, catalog number: 89850)
NaCl (Duchefa Biochemie, catalog number: S0520.5000)
RIPA (Cell nest, catalog number: CNR001-0100)
PierceTM BCA protein assay kit (Thermo Fisher Scientific, catalog number: 23225)
4× LDS sample buffer non-reducing (Thermo Fisher Scientific, 84788)
PLL solution (Sigma-Aldrich, catalog number: P4707)
Albumin (Biosesang, catalog number: AC1025-100-00)
Foil (SAM JIN FOIL, 30 cm × 30 m)
Triton X-100 (Sigma-Aldrich, catalog number: T9284)
Antifade-mounting media with DAPI (Vectashield, catalog number: H-1500)
Fractionation buffer (see Recipes)
SDS-detergent lysis buffer (see Recipes)
SDS-PAGE gel (see Recipes)
Part II. In vivo elimination of tauP301L neurofibrillary tangles
Injection syringes (Becton Dickinson Medical, catalog number: 1258696)
Dissection stainless steel tray (any brand)
Pins or tape (any brand)
Butterfly needles (Biosigma, catalog number: BSS250)
25- or 27-gauge infusion needle (KOREAVACCINE, catalog number: K201)
15 mL conical tube (Tarsons, catalog number: 546021)
1.5 mL e-tube (Axygen, catalog number: MCT-150-C)
24-well plate (Nest, catalog number: 702002, SPL, catalog number: 30024)
Kim-wipe or paper tower (Yuhan-Kimberly, catalog number: 41112)
Foil (SAM JIN FOIL, 30 cm × 30 m)
Microscope slide (Marienfield, catalog number: 1000612)
PBS (Biosesang, catalog number: P2007-1)
Avertin (Sigma-Aldrich, catalog number: T48402)
0.9% saline (Sigma-Aldrich, catalog number: 08059)
RIPA (Cell nest, catalog number: CNR001-0100)
Protease inhibitor (Abbkine, catalog number: BMP1001)
Phosphatase inhibitor (Sigma-Aldrich, catalog number: P0001)
Sucrose (Sigma-Aldrich, catalog number: S0389)
Sodium dodecyl sulfate (SDS) (Duchefa Biochemie, catalog number: S1377.1000)
Triple-distilled water (DIW)
4% paraformaldehyde (Biosesang, catalog number: pc2031-100-00)
Optional cutting temperature (OCT) compound (Sakura Tissue-Tek, catalog number: 4583)
Sodium azide (Thermo Fisher Scientific, catalog number: J21610-22)
Sudan black B (Sigma-Aldrich, catalog number: 199664)
Hoechst (Abcam, ab228550)
EtOH (DUKSAN, catalog number: 64-17-5)
0.01% PBST (see Recipes)
Blocking solution (see Recipes)
Injection drug (see Recipes)
Primary antibodies
Rabbit polyclonal anti-LC3 (Sigma-Aldrich, catalog number: L7543, 1:10,000 diluted in PBST solution)
Rabbit polyclonal anti-GAPDH (Bio World, catalog number: AP0063, 1:10,000 diluted in PBST solution)
Mouse monoclonal anti-Tau5 (Invitrogen, catalog number: AHB0042, 1:5,000 diluted in PBST solution)
Rabbit polyclonal anti-p-Tau (Invitrogen, catalog number: 44–752G, 1:5,000 diluted in PBST solution)
AT8 antibody (Abcam, catalog number: ab210703, 1:200 diluted in the blocking solution)
Secondary antibodies
Anti-rabbit IgG-HRP (Cell Signaling, catalog number: 7074, 1:10,000 diluted in PBST solution)
Anti-mouse IgG-HRP (Cell Signaling, catalog number: 7076, 1:10,000 diluted in PBST solution)
Cell
HeLa (ATCC, catalog number: CCL-2): the first immortal human cells to be grown in culture and the basis for countless scientific discoveries
SH-SY5Y-tauP301L-GFP (Innoprot): SH-SY5Y stable cells tagged with green fluorescence and expressing tau mutant [0N4R(P301L mutant)]
Mouse
TauP301L-BiFC transgenic mouse (Shin et al., 2020): the mouse model that has two non-fluorescent compartments (hTauP301L-VN173 and hTauP301L-VC155) turning on Venus fluorescence protein upon tau assembly when they are fused with TauP301L.
Plasmid
mRFP-GFP-hTauP301L plasmid [mRFP-GFP-LC3 plasmid (Cha-Molstad et al., 2016; Dalby et al., 2004) is modified with hTauP301L]
Equipment
Part I. In vitro degradation of tauP301L aggregates
Tabletop centrifuge (Eppendorf, model/catalog number: 5424R)
Sonicator (SONICS vibra cel)
Centrifuge (Hanil fieta 4)
37°C incubator (Hera cell vios 250i CO2 incubator)
Laser scanning confocal microscope (Zeiss 510 Meta)
Rocker, CR300 (Finepcr)
Part II. In vivo elimination of tauP301L neurofibrillary tangles
Dissecting forceps (JEUNG DO BIO&PLANT CO., catalog number: SD-S-04PK)
Tissue scissors (JEUNG DO BIO&PLANT CO., catalog number: S-49-16-SPK, S-54-10-SPK)
Vascular clamp (Surtex Instruments)
Micro spatula (Merck, catalog number: Z243213)
Peristaltic pump (Ismatec., catalog number: ISM834)
7 mL Dounce tissue grinder (Wheaton, catalog number: 35742)
Tissue grinder motor (Bel-Art, KA., catalog number: UB32-50)
Polypropylene pestle (Bel-Art, BA., catalog number: 19923-0001)
Rotor (FINEPCR, AG)
Tabletop centrifuge (Eppendorf, catalog number: 5424R)
-80°C deep freezer (Thermo Fisher Scientific, catalog number: TDE)
Cryostat (Leica, catalog number: CM3050S)
Cryomolds (Sakura Tissue-Tek, catalog number: 25608-922)
Paintbrush (AnB, catalog number: P000EACE)
ZEISS Axio Scan.Z1 (Carl Zeiss, Germany)
Software
LMS image software (Zeiss LSM Image Browser (ver. 4.2.0.121))
ImageJ (NIH, Bethesda)
Prism 6 software (GraphPad)
Procedure
Part I. In vitro degradation of tauP301L aggregates
Triton X-100-based insoluble/soluble fractionation assay
Plate SH-SY5Y-TauP301L-GFP cells at 1.2 × 106 cells/mL/well in a 6-well plate.
After 24 h, treat cells with DMSO or a combination of hydroxychloroquine (HCQ, 10 µM), okadaic acid (15 nM), or AUTOTAC degrader (100 nM) all together, at 37°C for 24 h.
Aspirate DMEM culture media, wash cells with 1 mL of PBS, and collect cells using 0.05% trypsin.
Collect trypsinized cells in a 1.5 mL e-tube. Centrifuge at 16,000 × g for 2 min to pellet the cells.
Re-suspend the cell pellet in 77 µL of fractionation buffer containing 0.5% Triton X-100 and incubate on ice for 30 min.
*Note: Vortex cells every 15 min.
Centrifuge at 16,000 × g for 10 min at 4°C.
Measure the concentration of proteins in the supernatant with a BCA.
Transfer 70 µL of the supernatant as the soluble fraction to a new e-tube, then boil with 30 µL of 5× Laemmli sample buffer at 100°C for 10 min.
Wash the pellet from Step A6 with PBS and centrifuge at 16,000 × g and 4°C. Repeat four times to obtain the insoluble fraction pellet.
Lyse the pellet from Step A6 with 70 µL of SDS-detergent lysis buffer and add 30 µL of 5× Laemmli sample buffer.
Boil at 100°C for 10 min and load on an SDS-PAGE gel.
Transfer onto polyvinylidene difluoride membrane at 100 V and 4°C for 2 h.
Block the membrane with 5% skim milk in PBS solution at room temperature (RT) for 30 min.
Incubate the membrane with indicated primary antibodies (anti-p-Tau, anti-Tau, anti-LC3, and anti-GAPDH) diluted in PBST solution on a rocker at 4°C overnight.
*Note: Information for primary antibodies is listed in Materials and Reagents.
Wash the membrane with 1× PBST for 10 min three times.
Incubate the membrane with host-specific HRP-conjugated secondary antibodies diluted in PBST solution on a rocker at 4°C for 1 h.
*Note: Information for secondary antibodies is listed in Materials and Reagents.
Repeat Step A15.
Detect the membrane with ECL solution using X-ray film (Figure 1).
Calculate autophagic flux (A.F.) indices based on the ratio of Tau and phospho-Tau (p-Tau) levels in the presence or absence of hydroxychloroquine (HCQ), and assess the changes in the indices upon AUTOTAC treatment.
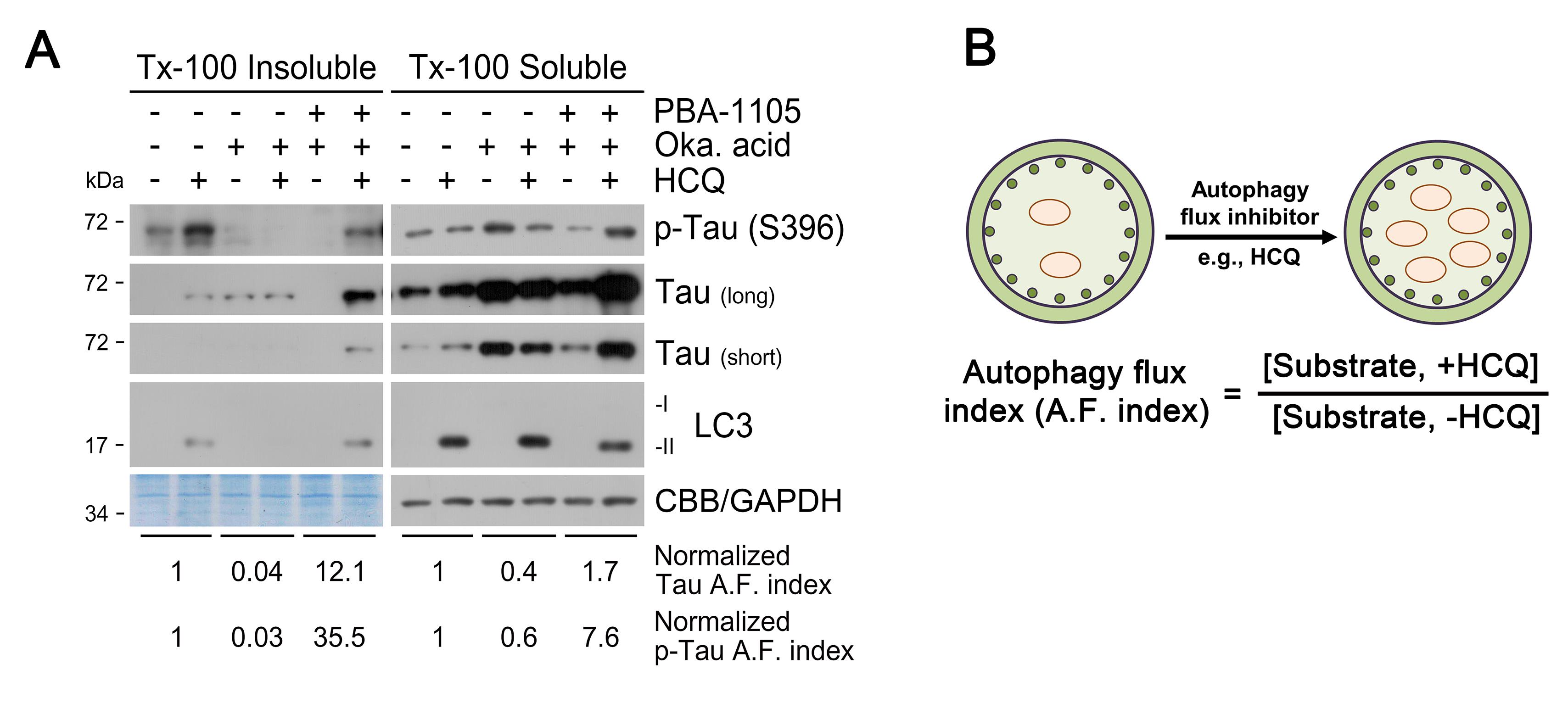
Figure 1. Triton X-100-fractionation assay. (A) SH-SY5Y-TauP301L-GFP cells were treated with the PBA-1105 AUTOTAC compound (100 nM, 24 h), okadaic acid (phosphatase inhibitor and tau hyperphosphorylation/aggregation inducer; 15 nM, 24 h), and/or HCQ (lysosome inhibitor; 10 µM, 24 h), and analyzed for immunoblotting (total tau was subjected to long or short exposure). Autophagic degradation of p-Tau/Tau is accelerated upon treatment with PBA-1105. (B) Normalized A.F. (autophagy flux) index indicates the relative ratio of the differences in the protein level between the absence and presence of HCQ.Oligomerization assay for detection and degradation of Tau aggregates
Seed SH-SY5Y-tauP301L-GFP cells at 1.2 × 106 cells/mL/well in 6-well plates.
Prepare PFF (Pre-formed Fibrill) solution at a concentration of 15 µL/well of 133.33 mM PFF suspended in a total of 200 µL of PFF buffer.
*Note: As an example, add 60 µL PFF to 140 µL of PFF buffer for a total of four wells.
Tap samples before sonication to facilitate pulsing.
Sonicate the samples with 10 pulses (1 pulse: 1 s on and 1 s off at 20% amplitude). Repeat three times, tapping samples after each pulse.
Add the sonicated PFF solution to a protein transfection reagent (PJ) vial.
*Note: One PJ vial could be used for up to two 6-well plates.
Incubate at RT for 5 min.
Add the PJ and the PFF mixture to 2 mL of serum-free RPMI media, mix well, and incubate with the cells.
After 4 h, add 200 µL of FBS to the media (final concentration: 10%) and incubate for an additional 20 h.
Alternatively, tau hyperphosphorylation and aggregation can be induced by treatment with the phosphatase inhibitor okadaic acid (15 nM, 24 h).
Add the AUTOTAC degrader (100 nM) to the media after 24 h of incubation of PFF or okadaic acid.
Using 0.05% trypsin, harvest cells by centrifugation at 16,000 × g for 2 min. Divide the mixture at a ratio of 1:9, re-suspend the smaller fraction with 50–100 µL of RIPA buffer, and incubate on ice for 30 min.
Centrifuge the smaller fraction suspension at 16,000 × g for 20 min, to obtain the supernatant.
Measure the concentration of proteins in the supernatants using the BCA.
Add 4× non-reducing LDS buffer to the larger fraction samples from Step B10, and heat at 95°C for 10 min.
Load 15–20 µg of larger fraction samples, according to the relative protein concentrations obtained in Step B12, and resolve the samples using a 4%–20% gradient SDS-PAGE.
*Note: Do not remove stacking gels when samples are transferred to membranes.
Immunoblot the samples using Tau5 (Total tau) or p-Tau (S396) antibodies (Figure 2).
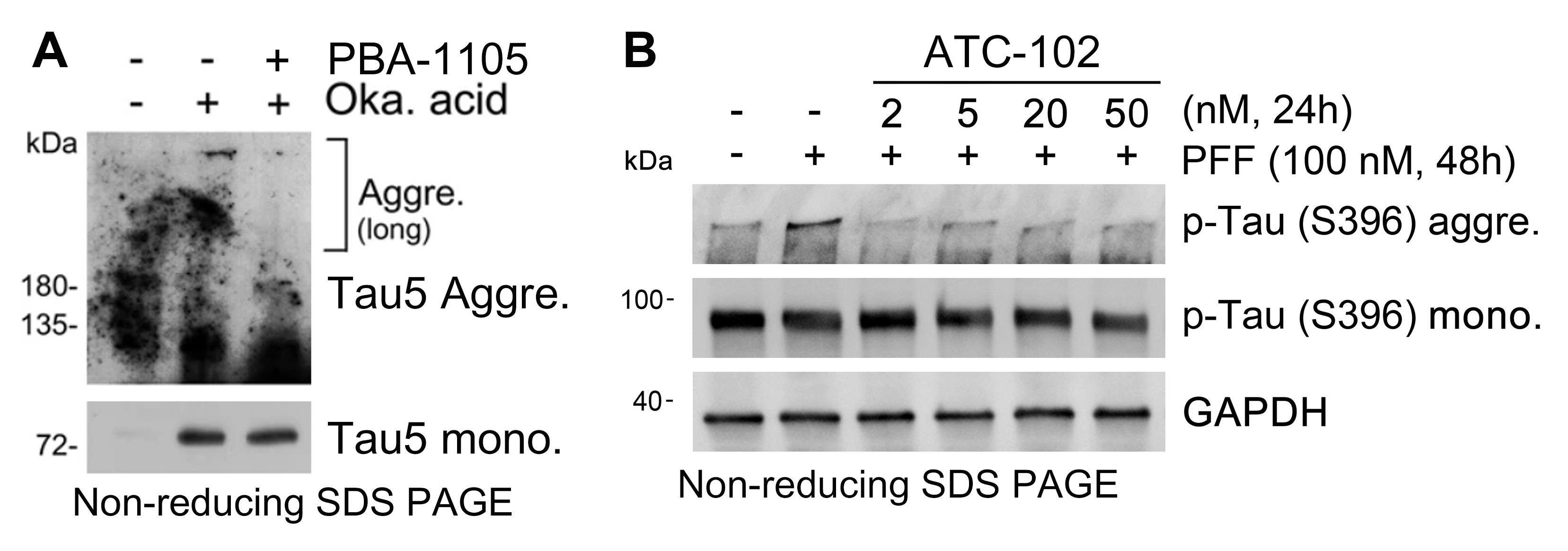
Figure 2. Detection of Tau aggregates with in vitro oligomerization assay. (A)Western blot of total tau monomer and aggregates in SH-SY5Y-tauP301L-GFP cells treated with okadaic acid (15 nM, 24 h) and the chemical chaperone-based PBA-1105 AUTOTAC compound (based on the FDA-approved 4-phenylbutyric acid; 100 nM, 24 h). (B) Western blot of pre-formed fibrils induced aggregates of p-Tau in SH-SY5Y-tauP301L-GFP cells treated with various concentrations of the oligomeric modulator-based ATC102 AUTOTAC compound (based on a protein oligomer/aggregate-specific warhead). AUTOTAC treatment via PBA-1105 or ATC-102 preferentially targets the aggregated, high-molecular weight species of mutant Tau.Lysosomal digestion assay of Tau aggregates
Seed HeLa cells at 5 × 104 cells/mL on coverslips coated with 10% poly-L-lysine (PLL), each placed in a 24-well plate.
*Note: Insert coverslips in each well of the plate, then incubate with 10% PLL in autoclaved triple-distilled water (DIW) for 30 min. Wash coverslips three times with DIW.
Transfect the mRFP-GFP-hTauP301L plasmid in HeLa cells with Lipo2000.
*Note: Incubate 0.5 µg/well of plasmid with 250 µL/well of opti-MEM, and 0.75 µL/well of Lipo2000 with 250 µL/well of opti-MEM for 5 min. Mix solutions and incubate for 15 min. Add the resulting mixture to cells (Fisher et al., 2018).
After 24 h of transfection, incubate cells in the presence or absence of okadaic acid (15 nM) and/or the AUTOTAC degrader (100 nM) for 24 h.
Remove the culture medium by aspiration and wash the cells with 1× PBS twice. A volume of 500 µL/well of 1× PBS is recommended for a 24-well plate.
*Note: Wrap the 24-well plate with aluminum foil and carry out the next steps in the dark whenever possible.
Fix cells with 4% paraformaldehyde (PFA) at RT for 15 min, followed by washing the coverslips three times on a rocking incubator with 1× PBS for 5 min per wash.
Permeabilize cells using 0.5% Triton X-100 solution at RT for 15 min, followed by washing identical to Step C5.
Mount coverslips with the antifade-mounting medium on the glass slide, and store at RT for 6 h for the medium to dry.
*Note: Avoid trapping air bubbles in the medium between the coverslip and the glass slide; if bubbles do form, press the coverslip only very slightly to remove them.
Obtain confocal images with a laser scanning confocal microscope. A magnification of 60× or greater is required to accurately observe intracellular puncta structures.
Repeat Step C8 as many times as required within three independent experiments, to count a minimum of 150 cells for statistical analysis.
To analyze lysosomal targeting of tau aggregates, analyze their acidification by counting the number of yellow (mRFP-EGFP) and red (mRFP-quenched EGFP) puncta structures, thus obtaining a GFP/RFP ratio (Kimura et al., 2007; Lopez et al., 2018).
*Note: Since GFP fluorescence is quenched in acidic pH, a decline in the GFP/RFP ratio means digestion of tau aggregates in autolysosomes (Figures 3, 4).
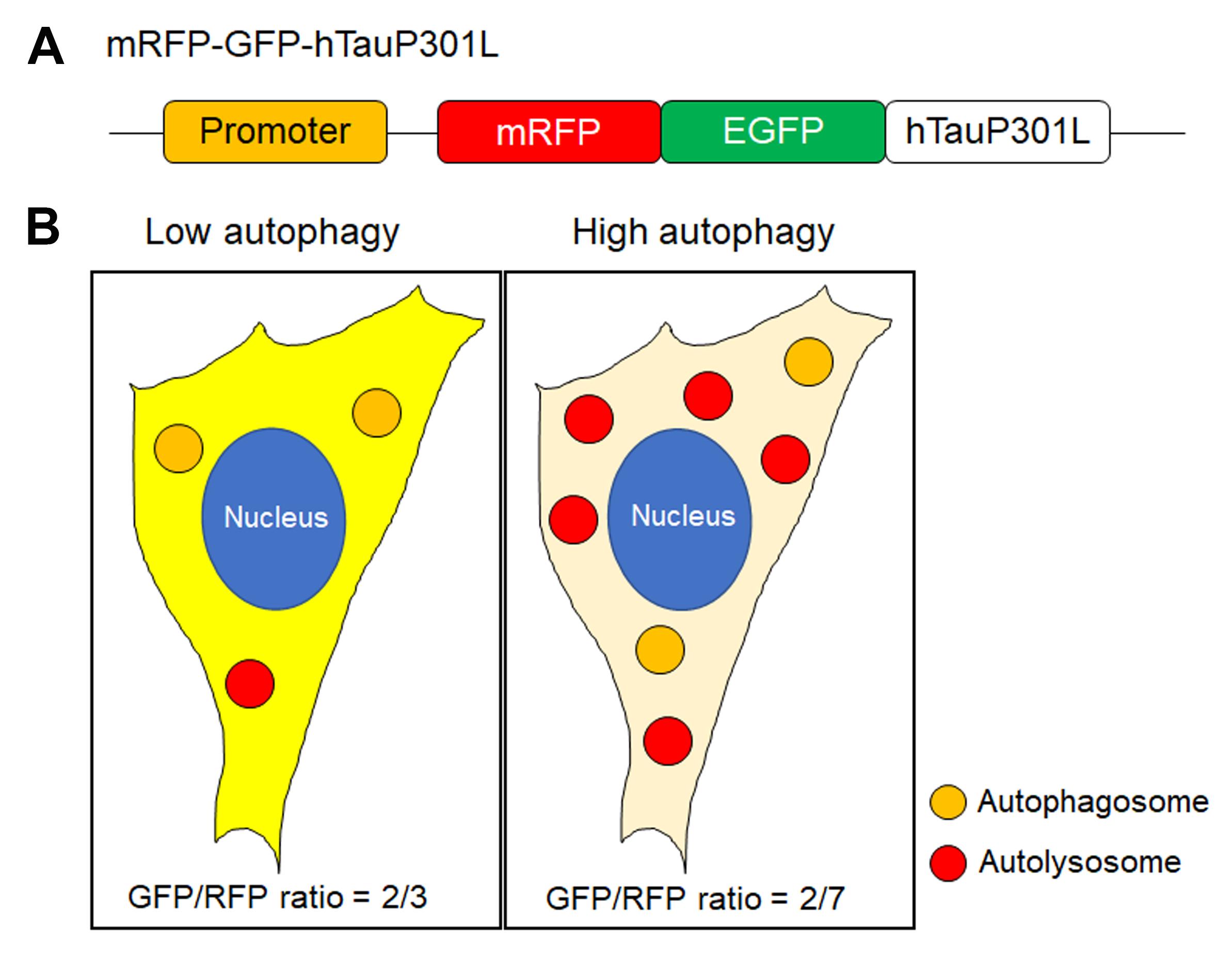
Figure 3. Measuring GFP/RFP ratio. (A)Schematic illustration of the design of mRFP-GFP-hTauP301L plasmid. (B) Description of measuring GFP/RFP ratio based on counting the puncta that are yellow (GFP and RFP) or red (RFP only). When autophagy is activated, the number of RFP only puncta is increased by quenching of GFP, which means lysosomal digestion.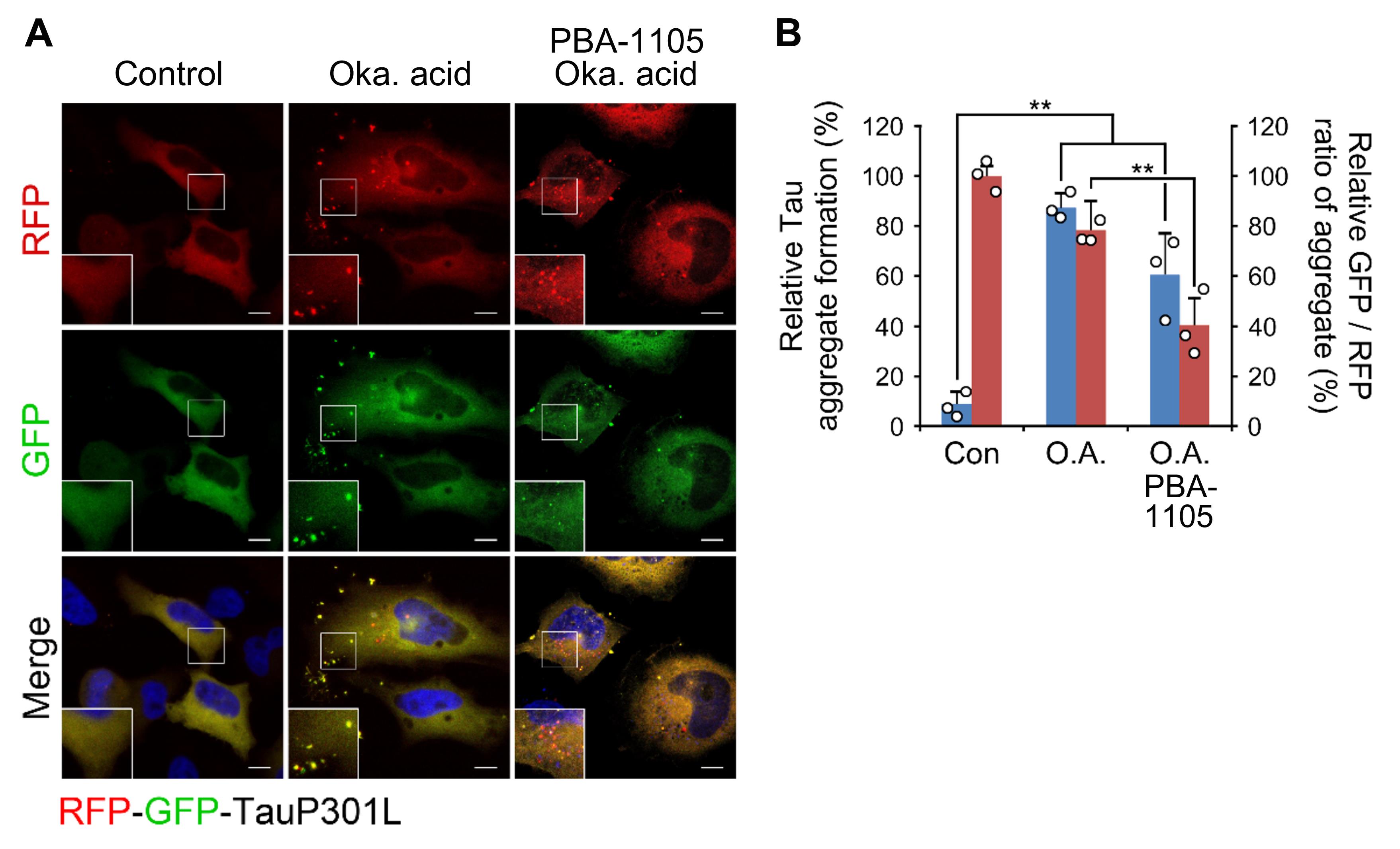
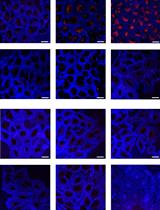
Figure 4. Examples of Tau aggregates lysosomal digestion assay. (A)Confocal image of mRFP-GFP-hTauP301L in HeLa cells, which are treated with okadaic acid (15 nM, 24 h) prior to the presence or absence of PBA-1105 (100 nM, 24 h). Scale bar, 10 µm. (B) Quantification of A for punctate formation (blue bars) and GFP/RFP ratio (red bars).
Part II. In vivo elimination of tauP301L neurofibrillary tangles
Intraperitoneal injection of AUTOTAC into the hTauP301L-BiFC tauopathy mouse model
To evaluate the effect of AUTOTAC degraders on the degradation of tau aggregates in vivo, inject PBA-1105 (20 or 50 mg/kg) dissolved in a 5% DMSO/10% solutol/85% PBS mixture, or PBS containing 30% polyethylene glycol (PEG) into the peritoneum of the hTauP301L-BiFC mouse (Shin et al., 2020), three times a week for four weeks (Figure 5).
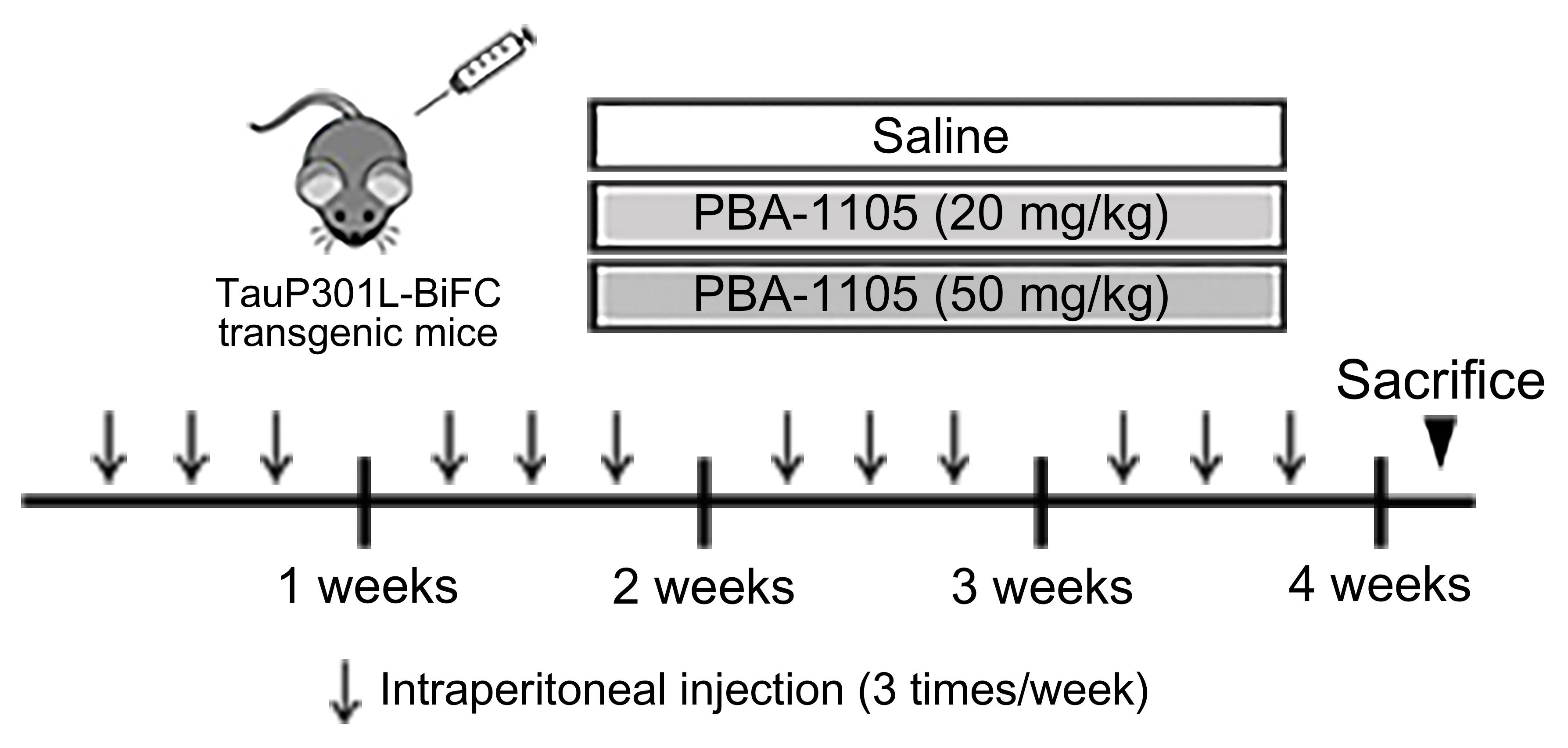
Figure 5. Injection timeline and details of PBA-1105 in hTauP301L-BiFC murine modelBrain tissue preparation
Anesthetize mice with an intraperitoneal injection of 2% avertin (2,2,2-Tribromoethanol), confirming full anesthesia via the paw pinch reflex.
Place mice on a dissection tray and fix each paw with pins or tapes. With forceps and scissors, open the skin and chest cavity, to expose the heart.
While holding the heart with forceps, insert the needle into the tip of the left ventricle. Then, cut the right atrium with scissors, and start the peristaltic pump (speed should be similar to the heart rate).
Perfuse with 0.9% saline, until the circulating fluid is clear and the liver turns white.
After perfusion, decapitate the mouse with scissors.
Cut the skin along the midline between the eyes of the mouse, and expose the skull.
Keep the end of the scissors on the foramen magnum and carefully cut off the midline of the skull towards the nose, to expose the brain.
Remove the skull covering the brain with forceps.
Gently transfer the brain to a 15 mL conical tube containing 1× PBS using a micro spatula.
RIPA-insoluble/soluble fractionation assay
Preparation of brain lysate (Figure 6)
*Note: Perform all steps on ice.
Place a homogenizer on ice to cool. Meanwhile, prepare the RIPA buffer mixture.
Place perfused mouse hemispheres in each 1.5 mL Eppendorf tube containing 500 µL of the RIPA buffer mixture.
Cut the brain tissue into small pieces with scissors.
Homogenize the samples with a tissue homogenizer, adding an additional 500 µL of the RIPA buffer mixture.
Incubate the samples on a rotor at 4°C for 2 h.
Centrifuge the samples at 14,000 × g and 4°C for 15 min.
Collect the supernatant (soluble fraction) in new 1.5 mL Eppendorf tubes and store in a -80°C deep freezer until use.
Wash the pellet from Step C1f with the washing solution to completely remove the soluble fractions.
Centrifuge at 14,000 × g and 4°C for 5 min. Repeat Steps C1h–i three times, but centrifuge at 14,000 × g and 4°C for 20 min for the final step.
Discard the supernatant.
Re-suspend pellet with 1 mL of the 2% SDS in triple-distilled water, containing 1% protease inhibitor and phosphatase inhibitor.
Incubate the samples at RT for 20 min.
Centrifuge the samples at 14,000 × g and 4°C for 10 min.
Collect the supernatant in a new 1.5 mL Eppendorf tube, to obtain the insoluble fraction.
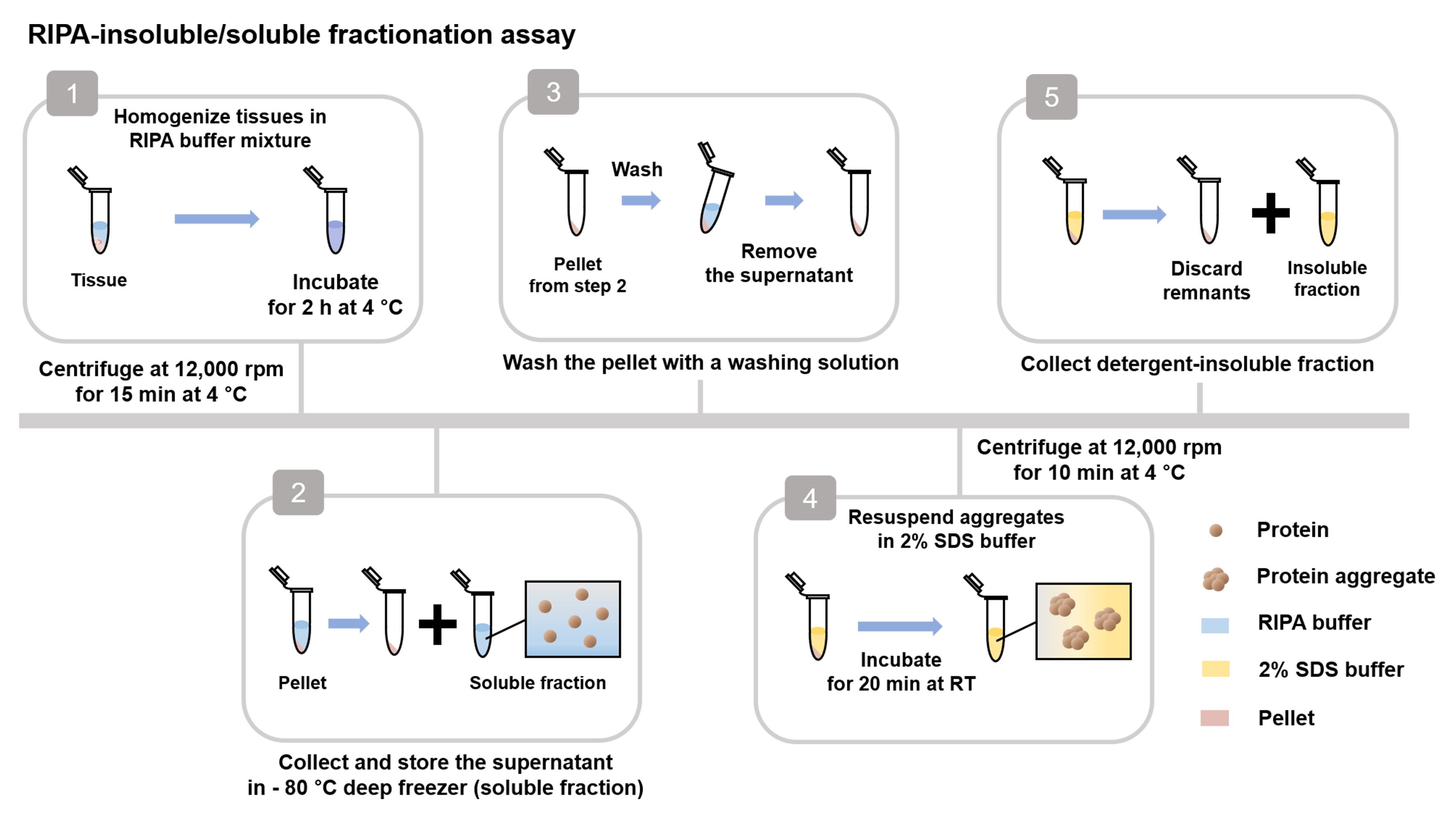
Figure 6. Workflow for RIPA-insoluble/soluble fractionation assayMeasure the concentration of proteins in the supernatant of Step C1g using the BCA and dilute proteins with RIPA buffer up to 1 µg/µL.
Transfer 70 µg of the soluble fraction supernatant to a new Eppendorf tube, add 30 µL of 5× Laemmli sample buffer, and boil at 100°C for 10 min.
Lyse the pellet from Step C1n with 70 µL of SDS-detergent lysis buffer and add 30 µL of 5× Laemmli sample buffer.
Boil at 100°C for 10 min and load on an SDS-PAGE gel.
Transfer onto a polyvinylidene difluoride membrane at 100 V and 4°C for 2 h.
Block the membrane with 5% skim milk in PBS solution at RT for 30 min.
Incubate the membrane with indicated primary antibodies (anti-Tau5 and anti-p-Tau) diluted in PBST solution on a rocker at 4°C overnight.
*Note: Information for primary antibodies is listed in Materials and Reagents.
Wash the membrane with 1× PBST for 10 min three times.
Incubate the membrane with host-specific HRP-conjugated secondary antibodies diluted in PBST solution on a rocker at 4°C for 1 h.
*Note: Information for secondary antibodies is listed in Materials and Reagents.
Repeat Step C9.
Detect the membrane with ECL solution using X-ray film (Figure 7).
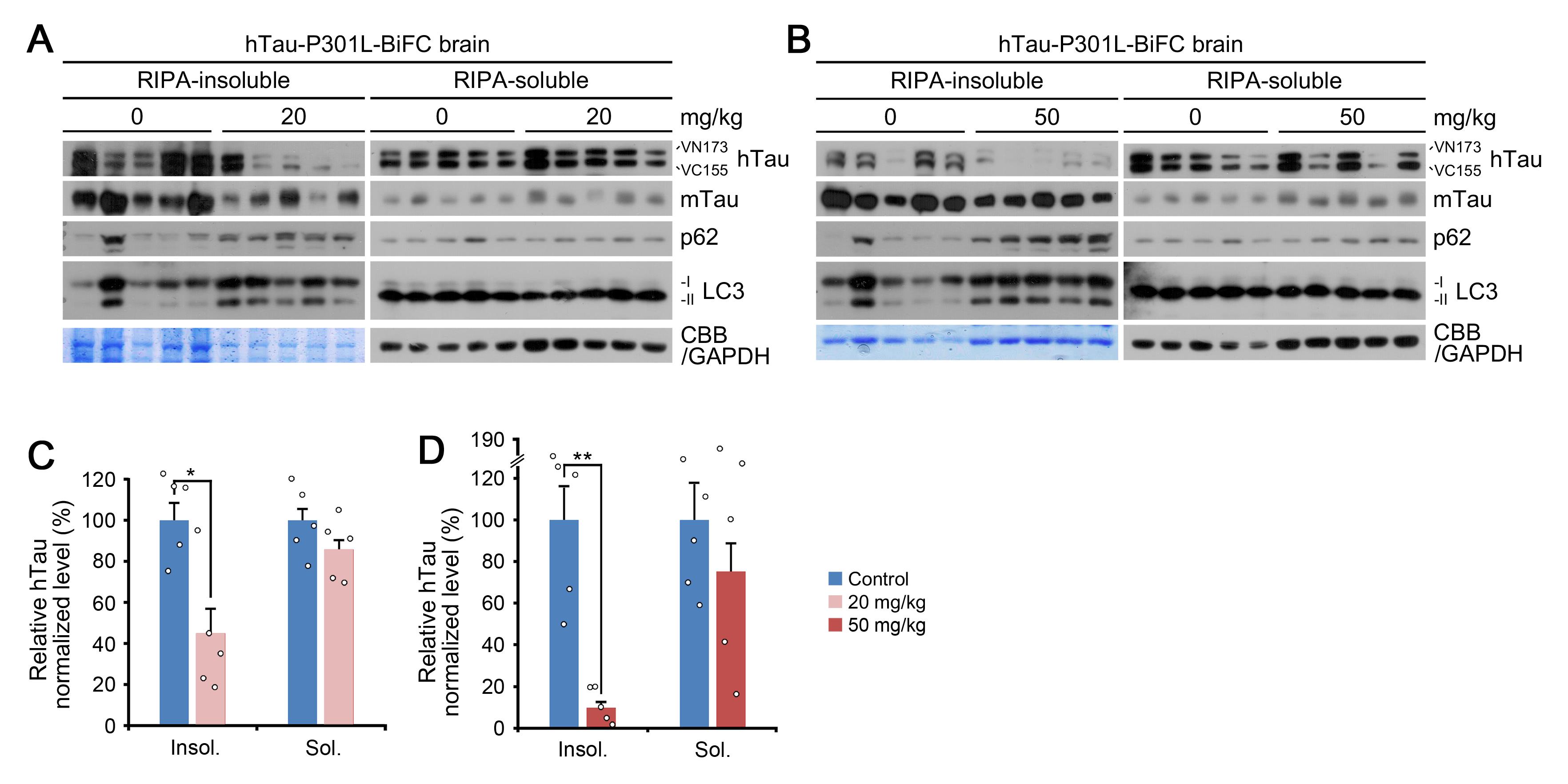
Figure 7. RIPA-insoluble/soluble fractionation assay. (A), (B) Fractionation assay in brain tissue of hTauP301L-BiFC mice intraperitoneally injected with PBA-1105 (20 or 50 mg/kg). (C), (D) Normalized densitometry of (A) and (B) for hTau levels, respectively (n = 5 mice). PBA-1105 AUTOTAC treatment selectively eliminates mutant human tau (hTau) in the RIPA-insoluble fraction, as opposed to murine tau (mTau) or RIPA-soluble human tau.Immunohistochemistry of hTauP301L-BiFC murine brain tissues
Mouse brain fixation
Rapidly fix the mouse hemisphere perfused with PBS containing 4% paraformaldehyde (PFA) at 4°C for 48 h.
Transfer the brains to 30% sucrose in PBS and confirm full immersion for cryoprotection.
Embedding in OCT compound
Before embedding, place cryomolds in the active cryostat with the temperature set to -20°C.
After cooling, label the samples on the surface of the cryomold with a permanent marker.
Remove excess liquid surrounding the brains by absorption with a Kim-wipe or paper towel, prior to freezing.
*Note: Excess liquid will form ice crystals on the surface of the brains and prevent the samples from properly adhering to the frozen OCT compound during embedding, interfering with sectioning.
Place a few drops of OCT onto the center of the bottom of the cryomold. Be careful to select the proper size embedding mold according to the size of the brains to be embedded.
Place the brain sample on several drops of the OCT compound and adjust its position. Gently push the brain with forceps to ensure that the bottom surface of the brain is placed properly, touching the face of the bottom, and the brain is located in the center of the mold.
Carefully add more OCT compound drop by drop onto the sample, until completely immersed.
*Note: Avoid the formation of air bubbles by removing them using forceps.
Place the OCT-covered sample within the cryomold in a cryostat, to harden the block.
Wrap the embedded block in foil and store in an -80°C deep freezer for storage.
Sectioning
Before sectioning, place paintbrushes, forceps, and cryostat block in the equipment for 20 min, to equilibrate to the chamber temperature of -20°C.
Prepare a 24-well plate and add 1× PBS to all wells.
Attach the sample to a circular cryostat block with a few drops of the OCT compound and equilibrate for approximately 10 min.
Cut sections 50–100 µm thick to trim the OCT compound surrounding the brain until the brain is visible and then set the dial to cut sections 30 µm thick.
*Note: Ensure sections are horizontally sliced as equally as possible.
Cut serially in 30 µm thickness coronal sections.
Use a paintbrush to gently collect the sections. Create series of sections representing the brain by distributing similar sections on subsequent wells (Figure 8).
*Note: Up to five sections can be added to each well.
*Note: For storage, transfer sections to 1× PBS solution containing 0.05% sodium azide at 4°C.
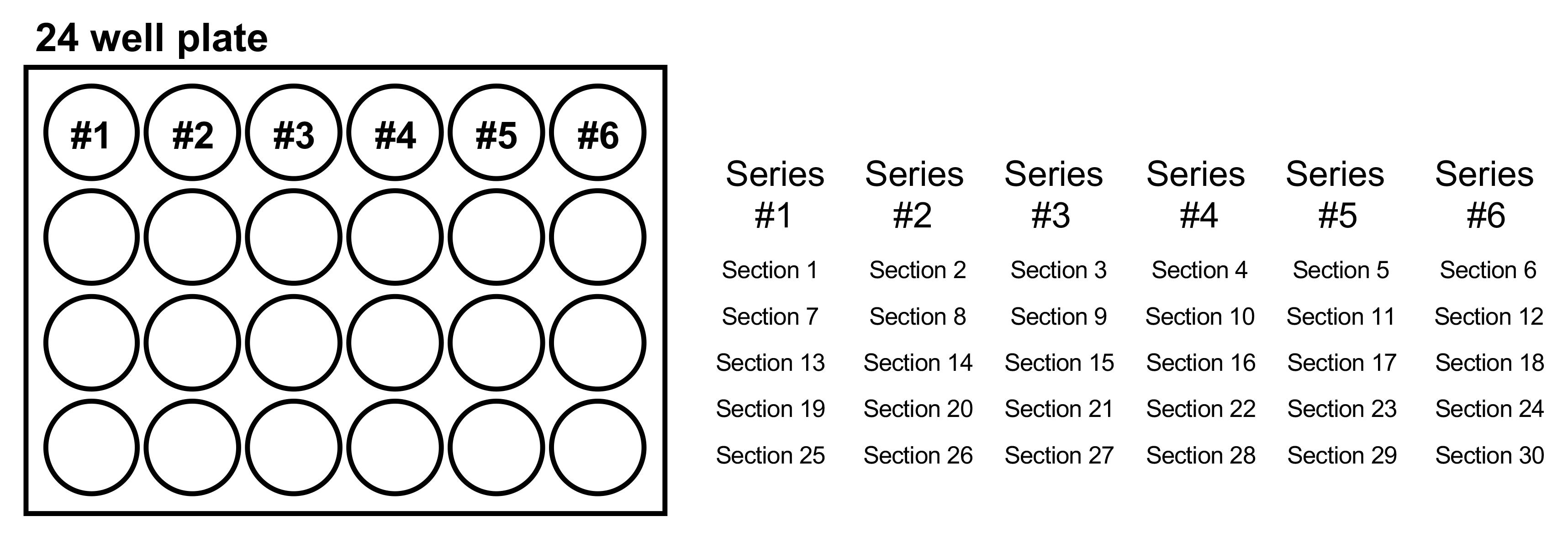
Figure 8. The manner of collecting brain sections. Each series includes five sections. As an example, sections 1, 7, 13, 19, and 25 are in Series #1. Series #2 includes sections 2, 8, 14, 20, and 26. Sections 3, 9, 15, 21, and 27 comprise series #3.Immunostaining: Sudan black B staining & AT8 antibody staining
*Note: Sudan black B staining reduces autofluorescence signals and improves the resolution of in situ hybridization-specific fluorescent signals in brain sections.
Stir 0.05% Sudan black B in 70% EtOH in the dark at RT for 2 h and let stand overnight (O/N). Alternatively, stir overnight and then filter with a 0.2 µm filter. This solution can be stored at 4°C for a week.
Remove the 1× PBS used in Step D3b by carefully aspirating it with a pipette tip, without touching the sections.
Wash with triple-distilled water for 5 min; repeat twice.
Immerse in the 0.05% Sudan black B solution for 10 min.
Wash with 0.1% PBST for 30 s; repeat three times.
Wash with triple-distilled water for 5 min.
Incubate the sections with 0.5 µg/mL Hoechst in triple-distilled water for 30 min.
Wash with triple-distilled water for 5 min; repeat twice.
Incubate the sections in blocking solution at 4°C O/N, by adding 1 mL to each well.
The following day, remove the blocking solution carefully, add 500 µL of anti-AT8 diluted 1:200 with the blocking solution, and incubate at 4°C O/N.
The next day, wash the sections with 1× PBS on a shaker at RT for 10 min each three times.
Prepare 500 µL of secondary antibody working solution by diluting 1:500 with blocking solution and incubate the sections on a shaker at 4°C O/N.
Wash the sections with 1× PBS, as described in Step D4k.
Carefully mount the sections on slide glasses using a paintbrush and dry at 4°C with a damp tissue. On the following day, dry them at RT, and then in the oven for 30 min.
*Note: When the sections move by a paintbrush, you have to be careful not to touch the sections, especially the targeting area.
BiFC fluorescence and AT8-stained signals of samples represent total and phosphorylated hTauP301L aggregates and neurofibrillary tangles, respectively.
Image acquisition and analysis (Figure 9)
Capture a region of interest using an automated fluorescence microscope system (ZEISS Axio Scan.Z1); λex = 460–490 nm and λex = 500–550 nm. Observe the total (BiFC signal) and phosphorylated (AT8 signal) tau within the hippocampus and cortex regions, specifically focusing on intracellular neurofibrillary tangles or aggregates.
For the quantitative analysis, fluorescence and total area of each puncta were measured using ImageJ. The significance of the data (p-value) was determined using two-tailed, unpaired Student’s t-test (degree of freedom = n-1) with Prism 6 software.
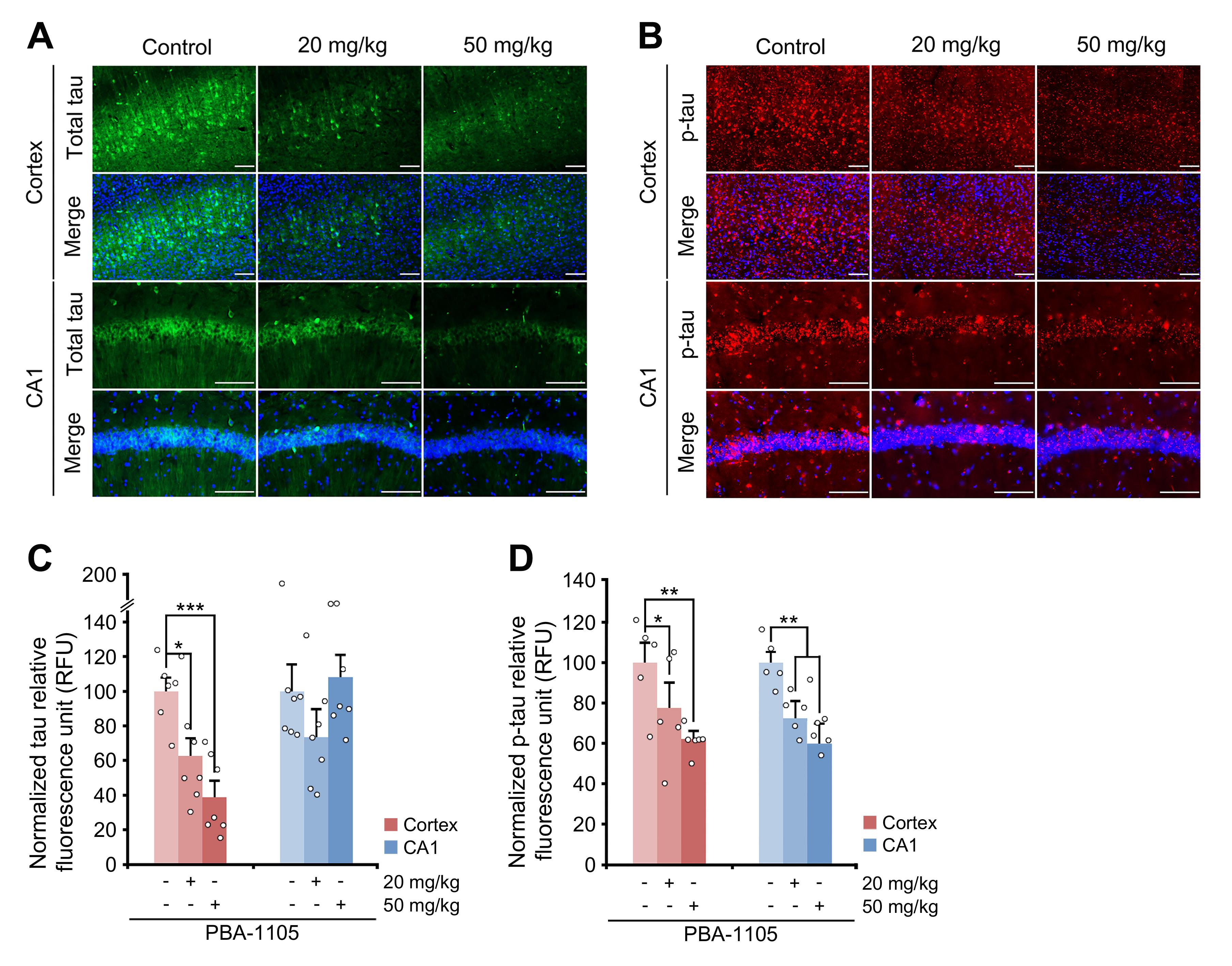
Figure 9. Immunohistochemistry of BiFC fluorescence. (A), (B) Immunohistochemistry of BiFC fluorescence for total hTau levels or AT8 fluorescence for total phosphorylated levels in hTauP301L-BiFC mice injected with PBA-1105, as outlined in Fig. 6. Scale bar, 100 μm. (C), (D) Quantification of BiFC or AT8 punctate fluorescence signals in (A) and (B), respectively (n = 7 mice).
Data analysis
For all data shown, stated values represent the mean ± S.D or S.E.M. of at least three independent experiments (unless otherwise stated). For each experiment, sample size (n) was determined as stated in the figure legends. For all experiments, p-values were determined using two-tailed, unpaired student’s test (degree of freedom = n-1) with Prism 6 software (GraphPad). Detailed information is provided in the original, open-access publication (https://doi.org/10.1038/s41467-022-28520-4).
Recipes
SDS-detergent lysis buffer
20 mM HEPES pH 7.9
0.2 M KCl
1 mM MgCl2
1 mM EGTA
1% Triton X-100
1% SDS
10% glycerol
Protease inhibitors
Phosphatase inhibitors
Fractionation buffer
100 mM NaCl
0.5 mM EDTA
20 mM Tris HCl pH 8
0.5% Triton X-100
SDS-PAGE Gel
30% acrylamide
1.5 M Tris-HCl pH 8.8
1 M Tris-HCl pH 6.8
10% SDS
10% APS
TEMED
2-Propanol
PFF buffer
10 mM HEPES
100 mM NaCl (1 M stock)
Distilled water
Injection drug
PBA-1105 (20 or 50 mg/kg)
5% DMSO/10% solutol/85% PBS mixture
30% polyethylene glycol (PEG)
1× PBS
RIPA buffer mixture
RIPA
1% protease inhibitor
1% phosphatase inhibitor
Washing solution
RIPA
1 M sucrose
1% protease inhibitor
1% phosphatase inhibitor
0.01% PBST
1× PBS
0.01% Tween
Blocking solution
1× PBS
0.3% Triton X-100
5% bovine serum albumin (BSA)
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (NRF-2020R1A5A1019023 and NRF-2021R1A2B5B03002614 to Y.T.K.), the Korea Health Technology R&D Project through the Health Industry Development Institute and Korea Dementia Research Center (KDRC) funded by the Ministry of Science, ICT, and Future Planning (MSIP) (HU21C0201 to C.H.J.), and the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2022R1A6A3A1307128711 to M.J.L. and 2022R1A6A3A13069514 to S.B.K.). This protocol was adapted from our previous work (Ji et al., 2022; https://doi.org/10.1038/s41467-022-28520-4)".
Competing interests
Ethics
Animal protocols followed the principles and practices outlined in the approved guidelines and also received ethical approval by the Institutional Animal Care and Use Committee (IACUC) of the Korea Institute of Science and Technology.
References
- Banik, S. M., Pedram, K., Wisnovsky, S., Ahn, G., Riley, N. M. and Bertozzi, C. R. (2020). Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature 584(7820): 291-297.
- Chamberlain, P. P. and Hamann, L. G. (2019). Development of targeted protein degradation therapeutics. Nat Chem Biol 15(10): 937-944.
- Cha-Molstad, H., Sung, K. S., Hwang, J., Kim, K. A., Yu, J. E., Yoo, Y. D., Jang, J. M., Han, D. H., Molstad, M., Kim, J. G., et al. (2015). Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat Cell Biol 17(7): 917-929.
- Cha-Molstad, H., Yu, J. E., Feng, Z., Lee, S. H., Kim, J. G., Yang, P., Han, B., Sung, K. W., Yoo, Y. D., Hwang, J., et al. (2017). p62/SQSTM1/Sequestosome-1 is an N-recognin of the N-end rule pathway which modulates autophagosome biogenesis. Nat Commun 8(1): 102.
- Dalby, B., Cates, S., Harris, A., Ohki, E. C., Tilkins, M. L., Price, P. J. and Ciccarone, V. C. (2004). Advanced transfection with Lipofectamine 2000 reagent: primary neurons, siRNA, and high-throughput applications. Methods 33(2): 95-103.
- Fisher, S. L. and Phillips, A. J. (2018). Targeted protein degradation and the enzymology of degraders. Curr Opin Chem Biol 44: 47-55.
- Heo, A. J., Kim, S. B., Ji, C. H., Han, D., Lee, S. J., Lee, S. H., Lee, M. J., Lee, J. S., Ciechanover, A., Kim, B. Y., et al. (2021). The N-terminal cysteine is a dual sensor of oxygen and oxidative stress. Proc Natl Acad Sci U S A 118(50): e2107993118.
- Heo, A. J., Ji, C. H. and Kwon, Y. T. (2022). The Cys/N-degron pathway in the ubiquitin-proteasome system and autophagy. Trends Cell Biol S0962-8924(22)00175-1.
- Ji, C. H., Kim, H. Y., Lee, M. J., Heo, A. J., Park, D. Y., Lim, S., Shin, S., Ganipisetti, S., Yang, W. S., Jung, C. A., et al. (2022). The AUTOTAC chemical biology platform for targeted protein degradation via the autophagy-lysosome system. Nat Commun 13(1): 904.
- Ji, C. H. and Kwon, Y. T. (2017). Crosstalk and Interplay between the Ubiquitin-Proteasome System and Autophagy. Mol Cells 40(7): 441-449.
- Ji, C. H., Kim, H. Y., Heo, A. J., Lee, S. H., Lee, M. J., Kim, S. B., Srinivasrao, G., Mun, S. R., Cha-Molstad, H., Ciechanover, A., et al. (2019). The N-Degron Pathway Mediates ER-phagy. Mol Cell 75(5): 1058-1072 e1059.
- Ji, C. H., Kim, H. Y., Heo, A. J., Lee, M. J., Park, D. Y., Kim, D. H., Kim, B. Y. and Kwon, Y. T. (2020). Regulation of reticulophagy by the N-degron pathway. Autophagy 16(2): 373-375.
- Ji, C. H., Lee, M. J., Kim, H. Y., Heo, A. J., Park, D. Y., Kim, Y. K., Kim, B. Y. and Kwon, Y. T. (2022). Targeted protein degradation via the autophagy-lysosome system: AUTOTAC (AUTOphagy-TArgeting Chimera). Autophagy 18(9): 2259-2262.
- Kimura, S., Noda, T. and Yoshimori, T. (2007). Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3(5): 452-460.
- Li, Z., Wang, C., Wang, Z., Zhu, C., Li, J., Sha, T., Ma, L., Gao, C., Yang, Y., Sun, Y., et al. (2019). Allele-selective lowering of mutant HTT protein by HTT-LC3 linker compounds. Nature 575(7781): 203-209.
- Lim, S., Kim, D., Ju, S., Shin, S., Cho, I. J., Park, S. H., Grailhe, R., Lee, C. and Kim, Y. K. (2018). Glioblastoma-secreted soluble CD44 activates tau pathology in the brain. Exp Mol Med 50(4): 1-11.
- Lopez, A., Fleming, A. and Rubinsztein, D. (2018). Seeing is believing: methods to monitor vertebrate autophagy in vivo. Open Biology 8: 180106.
- Moon, S. and Lee, B. H. (2018). Chemically Induced Cellular Proteolysis: An Emerging Therapeutic Strategy for Undruggable Targets. Mol Cells 41(11): 933-942.
- Peeraer, E., Bottelbergs, A., Van Kolen, K., Stancu, I. C., Vasconcelos, B., Mahieu, M., Duytschaever, H., Ver Donck, L., Torremans, A., Sluydts, E., et al. (2015). Intracerebral injection of preformed synthetic tau fibrils initiates widespread tauopathy and neuronal loss in the brains of tau transgenic mice. Neurobiol Dis 73: 83-95.
- Schapira, M., Calabrese, M. F., Bullock, A. N. and Crews, C. M. (2019). Targeted protein degradation: expanding the toolbox. Nat Rev Drug Discov 18(12): 949-963.
- Shin, S., Kim, D., Song, J. Y., Jeong, H., Hyeon, S. J., Kowall, N. W., Ryu, H., Pae, A. N., Lim, S. and Kim, Y. K. (2020). Visualization of soluble tau oligomers in TauP301L-BiFC transgenic mice demonstrates the progression of tauopathy. Prog Neurobiol 187: 101782.
- Thibaudeau, T. A., Anderson, R. T. and Smith, D. M. (2018). A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat Commun 9(1): 1097.
- Takalo, M., Salminen, A., Soininen, H., Hiltunen, M. and Haapasalo, A. (2013). Protein aggregation and degradation mechanisms in neurodegenerative diseases. Am J Neurodegener Dis 2(1): 1-14.
- Takahashi, D., Moriyama, J., Nakamura, T., Miki, E., Takahashi, E., Sato, A., Akaike, T., Itto-Nakama, K. and Arimoto, H. (2019). AUTACs: Cargo-Specific Degraders Using Selective Autophagy. Mol Cell 76(5): 797-810 e710.
- Yoo, Y. D., Mun, S. R., Ji, C. H., Sung, K. W., Kang, K. Y., Heo, A. J., Lee, S. H., An, J. Y., Hwang, J., Xie, X.-Q., et al. (2018). N-terminal arginylation generates a bimodal degron that modulates autophagic proteolysis. Proc Natl Acad Sci U S A 115(12): E2716-E2724.
- Zhang, Y., Mun, S. R., Linares, J. F., Ahn, J. W., Towers, C. G., Ji, C. H., Fitzwalter, B. E., Holden, M. R., Mi, W., Shi, X., et al. (2018). ZZ-dependent regulation of p62/SQSTM1 in autophagy. Nat Commun 9(1): 1-11.
Article Information
Copyright
© 2023 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lee, M. J., Kim, S. B., Kim, H. Y., Lee, S. J., Lee, J. S., Kwon, Y. T. and Ji, C. H. (2023). Methods to Detect AUTOphagy-Targeting Chimera (AUTOTAC)-mediated Targeted Protein Degradation in Tauopathies. Bio-protocol 13(2): e4594. DOI: 10.21769/BioProtoc.4594.
Category
Drug Discovery > Drug Screening
Neuroscience > Nervous system disorders > Cellular mechanisms
Cell Biology > Cell-based analysis > Autophagic activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








