- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Faster Bacterial Gene Cloning Using the Brick into the Gateway (BiG) Protocol
(*contributed equally to this work) Published: Vol 12, Iss 24, Dec 20, 2022 DOI: 10.21769/BioProtoc.4576 Views: 2348
Reviewed by: Alba BlesaDemosthenis ChronisChangyi Zhang

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
TUNEL Assay to Assess Extent of DNA Fragmentation and Programmed Cell Death in Root Cells under Various Stress Conditions
Amit K. Tripathi [...] Sneh Lata Singla-Pareek
Aug 20, 2017 15020 Views
Efficient Transient Gene Knock-down in Tobacco Plants Using Carbon Nanocarriers
Gozde S. Demirer and Markita P. Landry
Jan 5, 2021 6378 Views
Abstract
Cloning systems like Gateway and Golden Gate/Braid are known because of their efficiency and accuracy. While the main drawback of Gateway is the expensive cost of the enzymes used in its two-step (LR and BP) reaction, Golden Gate requires non-reusable components due to their specific restriction sites. We present the Brick into the Gateway (BiG) protocol as a new cloning strategy, faster and more economic method that combines (i) reusable modules or bricks assembled by the GoldenBraid approach, and (ii) Gateway LR reactions [recombination of attachment sites: attL (L from left) and attR (R from right)] avoiding the BP reaction [recombination of attachment sites: attP (P from phage) and attB (B from bacteria)] usually necessary in the Gateway cloning. The starting point is to perform a PCR reaction to add type IIS restriction sites into DNA fragments generating specific fusion sites. Then, this PCR product is used to design GoldenBraid bricks, including the attL Gateway recombination sites. Using the Golden Gate method, these bricks are assembled to produce an attL1–gene of interest–attL2 fragment, which is integrated into a compatible vector producing a Gateway entry vector. Finally, the fragment containing the target gene is recombined by LR reaction into the Gateway destination vector.
Graphical abstract

Background
DNA cloning tools have been improved to optimize the efficiency of cloning systems. New cloning methods were designed, such as (i) Gibson assembly, which integrates a number of DNA fragments in a single reaction using a mix of the three enzymes exonuclease, DNA polymerase, and DNA ligase (Gibson et al., 2009), (ii) Gateway, which requires specific recombination sites (atts) in both the DNA fragment and plasmids used in the two reactions steps: BP reaction—in which an entry vector, harboring a DNA fragment flanked by two attL sites, is created; and LR reaction—wherein an expression vector containing a DNA fragment flanked by attB sites is generated (Karimi et al., 2007; Invitrogen, USA); (iii) Golden Gate cloning, a method based on the use of type IIS endonucleases that have cut sites distal from their recognition sites producing overhangs; the complementary overhangs are traditionally ligated by T4 DNA ligase (Engler et al., 2008); (iv) USER cloning (Uracil-Specific Excision Reagent), which employs primers containing uracil to perform PCR reactions; the PCR product is incubated with a commercial deoxyuridine-excision enzyme to eliminate the uracil and generate 3' overhangs that allow the direct cloning of PCR fragments into a USER-compatible vector (Geu-Flores et al., 2007; Hansen et al., 2014); and (v) In-Fusion, in which four or more DNA fragments containing an overlapping of 15 bp at both ends are combined by a reaction equivalent to recombination; the ends overlap is achieved by including the 15 bp in the primers used for DNA amplification (Zhu et al., 2007).
Gateway and Golden Gate are the most used cloning systems. Both have high efficiency and accuracy. However, the drawback of Gateway is the dependency on specific commercial enzymes used for the two clonase reactions, BP and LR, which makes this technique more expensive. On the other hand, Golden Gate requires specific restriction sites in the vector as well as in the DNA fragments, which could be a limiting factor when designing constructs for a large number of genes, as the components of this method are generally not reusable (Marillonnet and Grützner, 2020). Thus, modular assembly systems have been developed to standardize the parts, or bricks (DNA fragments of interest), which is crucial for cloning construction. These modular systems facilitate cloning assembly, making it simpler, more versatile, and autonomous (Patron et al., 2015).
Here, we provide a new cloning method, Brick into the Gateway (BiG), that uses the GoldenBraid/Gate methods to assemble the bricks and the LR reaction from the Gateway system to transfer DNA fragments into any destination vector. Using the GoldenBraid system, we generated standard bricks of the attL sites into an acceptor plasmid pUPD2, which can be combined with other bricks harboring any DNA fragments through a Golden Gate reaction. After this reaction, an entry vector containing the DNA of interest flanked by attL sites is generated, allowing its transfer to a destination vector using Gateway LR clonase. BiG is a low-cost method with a short processing time and directional cloning, whose efficiency was successfully demonstrated (Ferigolo et al., 2022). Most importantly, BiG allows the use of assembled bricks in different studies due to its common syntax (Ferigolo et al., 2022).
Materials and Reagents
Enzymes
High-fidelity DNA polymerase with proper buffer (e.g., platinum SuperFi II polymerase, InvitrogenTM, catalog number: 12361010)
Restriction enzyme cloning BsmBI-v2 with proper buffer (New England Biolabs Inc., catalog number: R0580)
Restriction enzyme cloning BsaI-HFv2 with proper buffer (New England Biolabs Inc., catalog number: R3733)
T4 DNA ligase with proper buffer (e.g., Thermo Fisher ScientificTM, catalog number: EL0014)
Gateway LR clonase II enzyme mix with proper buffer (InvitrogenTM, catalog number: 11791100)
GoTaq green master mix (PromegaTM, catalog number: M7122)
Cloning vectors
Vector harboring attL sites (e.g., pENTRTM/D-TOPOTM)
LM22_attL1 (Addgene, catalog number: 184008)
LM22_attL2 (Addgene, catalog number: 184010)
Universal GoldenBraid acceptor (e.g., pUPD2)
Golden Gate acceptor vector (e.g., pICSL86900OD)
Gateway expression vector of choice
Kits
Gel extraction kit (e.g., QIAEX II, QIAGENTM, catalog number: 20021)
Plasmid Miniprep kit (e.g., GeneJET, Thermo Fisher ScientificTM, catalog number: K0702)
Others
dNTPs, 10 mM (Thermo Fisher ScientificTM, catalog number: R0192)
PCR primers
DNase- and RNase-free water
PCR tubes, 0.2 mL (AxygenTM, catalog number: 14-222-262)
6×loading dye (New England Biolabs Inc., catalog number: B7024S)
1 Kb plus DNA ladder (e.g., InvitrogenTM, catalog number: 10787018)
Agarose 1.5% gel (BioBasic, catalog number: AB0014)
SYBRTM Safe DNA gel stain (InvitrogenTM, catalog number: S33102)
Escherichia coli DH10α chemically competent cells (Thermo Fisher Scientific, catalog number: 18265017)
LB plates with the appropriate antibiotic, IPTG (Fermentas, catalog number: R0392), and X-gal substrate (Fermentas, catalog number: R0401)
Chloramphenicol (Sigma-Aldrich, catalog number: C0378)
Esp31 (Thermo Fisher ScientificTM, catalog number: ER0451)
Eco31I (Thermo Fisher ScientificTM, catalog number: ER0291)
GoldenBraid (GB) reaction (TV = 10 µL) (see Recipes)
LB medium (see Recipes)
Golden Gate reactions (TV = 10 µL) (see Recipes)
LR reaction (TV = 5 µL) (see Recipes)
Equipment
PCR cycler (Mastercycler® nexus, catalog number: 6330000021)
Microwave oven
Gel electrophoresis chamber
Blue-Light transilluminator (Safe ImagerTM 2.0, catalog number: G6600)
NanoDropTM OneC Microvolume UV-Vis spectrophotometer (Thermo Fisher ScientificTM, catalog number: 13-400-519)
Heat block at 42 °C
ColiRollersTM plating beads, Novagen® (Sigma-AldrichTM, catalog number: 71013-3)
Incubators at 37 °C
Software
Benchling R&D Cloud (https://www.benchling.com/)
Procedure
Primers design
Primers should be designed according to the GoldenBraid syntax, including BsmBI (Esp3I) restriction sites followed by pUPD2 and GoldenBraid fusion sites in both extremities (Figure 1A).
In case the sequence from the gene or DNA fragment of interest presents BsmBI (Esp31) or BsaI restriction sites, the fragment should be divided into sub-fragments, inserting synonymous mutations in the primers to silence the restriction site (Patron et al., 2015).
Alternatively, the target gene can be amplified using primers with the BsaI sites instead of BsmBI sites, with the forward overhang GGGGTCTCAAATG and reverse overhang GGGGTCTCAAAGC (Figure 1B). The PCR products can be used directly in the Golden Gate reaction without the pUPD2 cloning step.

Figure 1. Primer design to clone attL sequences and the target gene into GoldenBraid bricks, according to Vazquez-Vilar et al. (2017). Primers are designed to use the standard (A) or alternative (B) BiG protocol. Green box: BsmBI recognition site with the pUPD2 fusion sites (underlined); Orange box: fusion site to follow the Golden Gate assembly syntax; Purple box: specific sequence to amplify attL or target gene; Blue box: BsaI recognition site; RC: reverse complement.GoldenBraid bricks assembly into pUPD2 vector
Individually amplify attL1 and attL2 fragments, as well as the target gene, by touchdown PCR using a high-fidelity DNA polymerase such as the platinum SuperFi II polymerase.
Use a pENTRTM/D-TOPOTM vector as a DNA template to amplify attL1 and attL2 sequences.
Perform the PCR reaction in 50 μL of final volume according to the enzyme manufacturer's instructions.
Touchdown PCR steps: 98 °C for 1 min, followed by nine cycles of 98 °C for 30 s, proper melting temperature for 30 s, and 65 °C for 1 min, starting with 66 °C of melting temperature and decreasing 1 °C each cycle, plus 30 cycles of 98 °C for 30 s, 54 °C for 30 s, and 65 °C for 1 min, and ending with 65 °C for 3 min.
Add loading dye to the PCR samples to the final concentration of 1× and perform the electrophoresis using the total PCR reaction volume and the 1 Kb plus DNA ladder in 1.5% agarose with 0.002% of SYBRTM Safe DNA gel stain.
Confirm the PCR amplification and the amplicon size, 150 pb, by visualizing in the Blue-Light transilluminator.
Cut the DNA band corresponding to attLs or the target gene fragments and purify it from the agarose using the gel extraction kit.
Note: Purification can be performed directly from the PRC products.
Quantify DNA concentration using the spectrophotometer.
Clone the attL1, attL2, and target gene fragments into the GoldenBraid acceptor vector (e.g., pUPD2) by an assembly reaction using BsmBI restriction enzyme and T4 DNA ligase (Figure 2).
Set up the GoldenBraid reactions according to Recipe 1 (see Recipes).
Thermal cycler steps: 37 °C for 10 min followed by 30 cycles of 37 °C for 3 min and 25 °C for 4 min, ending with 37 °C for 10 min and 80 °C for 5 min.
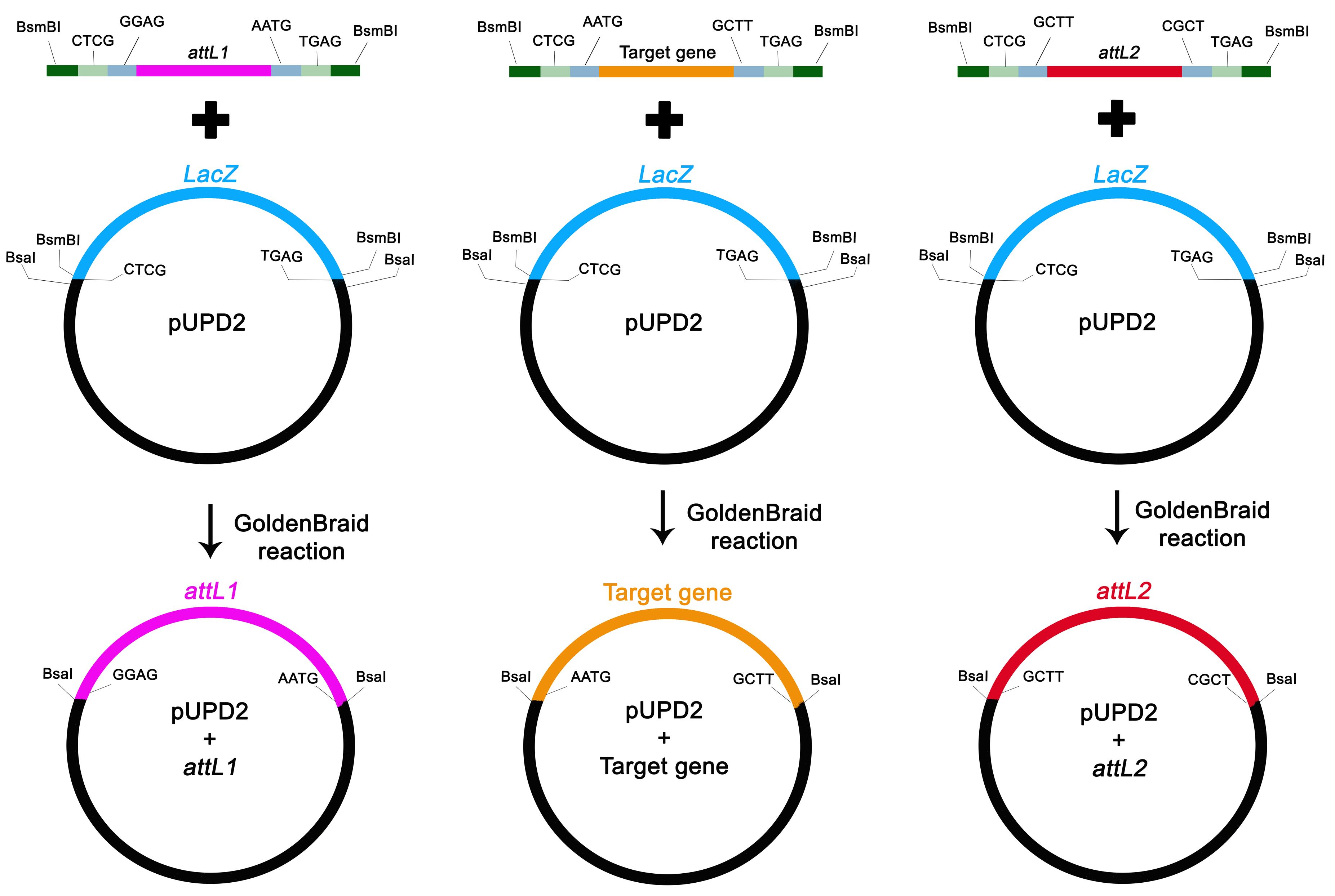
Figure 2. Schematic GoldenBraid bricks assembly. attL s and target gene individual PCR fragments (top) flanked by the BsmBI (dark green box), pUPD2 fusion sites (light green box), and Golden Gate fusion sites (light blue box) along with the pUPD2 vector (middle), were submitted to the GoldenBraid reaction to obtain the first bricks (bottom). All fusion sites are in the sense direction.Transform E. coli DH10α chemically competent cells with the total assembling reaction volume by heat shock.
Plate the cells using plating beads on LB (see Recipe 2) with chloramphenicol (25 μL/mL), IPTG (1 mM), and X-gal (200 μg/mL) and incubate cell plates at 37 °C overnight.
Select from two to five isolated white colonies and pick them to perform PCR colony using GoTaq green master mix to confirm the assembly construction.
Perform PCR colony using specific primers for each cloned DNA sequence.
Inoculate a single positive colony in liquid LB medium with chloramphenicol (25 μL/mL) and incubate shaking at 37 °C overnight.
Extract the plasmid DNA from the liquid culture using plasmid Miniprep kit and quantify DNA concentration with the spectrophotometer.
Golden Gate assembly
Perform the Golden Gate (GG) assembly with attL1-pUPD2, attL2-pUPD2, target gene-pUPD2, and a GG acceptor vector (e.g., pICSL86900OD) using BsaI restriction enzyme and T4 DNA ligase (Figure 3).
Set up the Golden Gate reactions according to Recipe 3 (see Recipes).
Thermal cycler steps: 37 °C for 10 min followed by 30 cycles of 37 °C for 3 min and 25 °C for 4 min, ending with 37 °C for 10 min and 80 °C for 5 min.
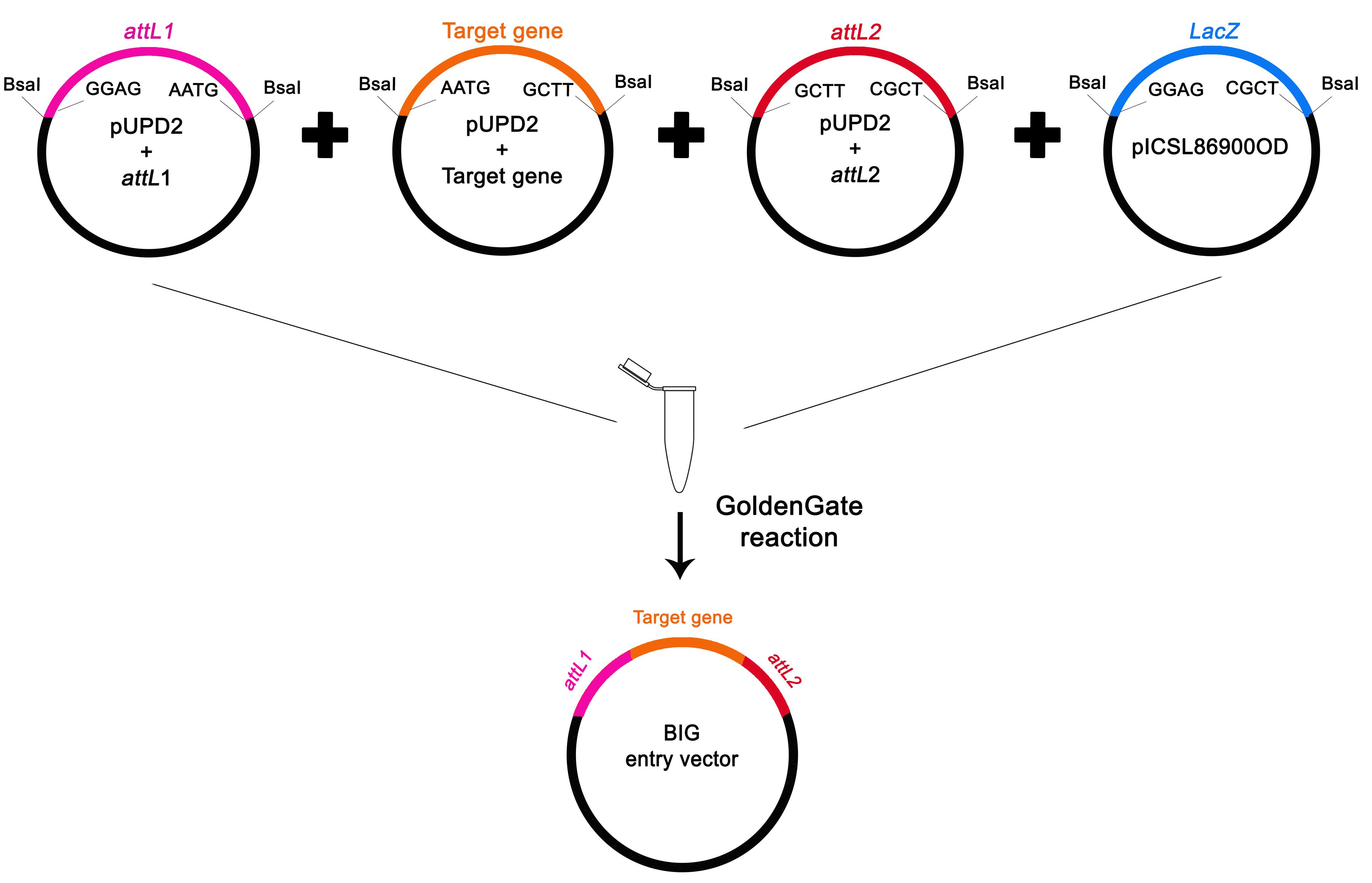
Figure 3. Schematic Golden Gate assembly to produce the BiG entry vector. Individual bricks harboring attL s and the target gene were submitted to the Golden Gate reaction with the pICSL86900OD vector.Transform E. coli DH10α chemically competent cells with the total assembly reaction volume and plate on selective LB medium with the proper antibiotic as described previously.
Select from two to five isolated white colonies by performing PCR colony. Then, one colony can be used to cultivate cells in a liquid LB medium following the plasmidial DNA extraction.
Perform PCR colony using specific primers for each cloned DNA sequence.
LR reaction
Perform LR reaction using the GoldenGate entry vector containing the gene of interest flanked by attL sites and the Gateway expression vector of choice.
To reduce costs, we performed the LR reaction with half the volume indicated by the manufacturer’s protocol in an overnight incubation at room temperature (see Recipe 4).
Transform E. coli DH10α chemically competent cells with the LR reaction and plate in selective LB medium with the proper antibiotic as described previously.
Plasmidial DNA extraction from positive colonies selected by PCR colony can be used in further experiments.
Data analysis
To confirm the nucleotide sequence and the correct assembly of the DNA constructions, miniprep samples can be sent for Sanger sequencing.
Notes
E. coli liquid culture expressing the attL-pUPD2 GoldenBraid bricks can be used to prepare a glycerol stock and stored at -80 °C for future assemblies with different target genes. Furthermore, these vectors are available in the plasmid repository Addgene as LM22_attL1 (catalog number: 184008) and LM22_attL2 (catalog number: 184010).
BsmBI restriction enzyme can be substituted for Esp31 (Thermo Fisher ScientificTM, catalog number: ER0451).
BsaI restriction enzyme can be substituted for Eco31I (Thermo Fisher ScientificTM, catalog number: ER0291).
The Golden Gate fusion sites were defined according to the GoldenBraid Standard (Vazquez-Vilar et al., 2017). However, different fusion sites can be used.
Recipes
GoldenBraid (GB) reaction (TV = 10 µL)
Reagent Final concentration Amount T4 DNA ligase buffer (10×) 1× 1.0 µL Restriction enzyme-specific buffer (10×) 1× 1.0 µL T4 DNA ligase -- 1.0 µL BsmBI restriction enzyme -- 1.0 µL GB acceptor vector (e.g., pUPD2) 10 ng µL-1 100 ng DNA fragment 5–10 ng µL-1 50–100 ng DNase- and RNase-free water -- q.s. 10 µL LB medium (TV = 1L)
Reagent Final concentration Amount Tryptone -- 10 g Yeast extract -- 5 g NaCl 1 M 5 g For a solid medium add agar at a final concentration of 8%.
Golden Gate (GG) reactions (TV = 10 µL)
Reagent Final concentration Amount T4 DNA ligase buffer (10×) 1× 1.0 µL Restriction enzyme–specific buffer (10×) 1× 1.0 µL T4 DNA ligase -- 1.0 µL BsaI restriction enzyme -- 1.0 µL GG acceptor vector (e.g., pICSL86900OD) 15 ng µL-1 150 ng attL1-pUPD2 5–10 ng µL-1 50–100 ng attL2-pUPD2 5–10 ng µL-1 50–100 ng target gene-pUPD2 5–10 ng µL-1 50–100 ng DNase- and RNase-free water -- q.s. 10 µL LR reaction (TV = 5 µL)
Reagent Final concentration Amount LR clonase II Mix (5×) 1× 1.0 µL Gateway expression vector 15 ng µL-1 150 ng BIG entry vector 15 ng µL-1 150 ng Tris-EDTA (TE) buffer pH 8.0 -- q.s. 5 µL 1Overnight incubation at room temperature.
Acknowledgments
This work was supported by The Sao Paulo Research Foundation – FAPESP (2018/17441–3, 2019/20157–8, 2020/12940-1, 2021/14640-8), and the Coordination for the Improvement of Higher Education Personnel – CAPES (88882.461731/2019-01; 88887.309513/2018-00, and 88887.613665/2021-00). This protocol was adapted from our recent work (Ferigolo et al., 2022).
Competing interests
The authors declare non-financial competing interests.
References
- Engler, C., Kandzia, R. and Marillonnet, S. (2008). A one pot, one step, precision cloning method with high throughput capability. PLoS One 3(11): e3647.
- Ferigolo, L. F., Vicente, M. H. and Nogueira, F. T. S. (2022). Brick into the Gateway (BiG): A novel approach for faster cloning combining Golden Gate and Gateway methods. Plasmid 121: 102630.
- Gibson, D. G., Young, L., Chuang, R. Y., Venter, J. C., Hutchison, C. A., 3rd and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6(5): 343-345.
- Geu-Flores, F., Nour-Eldin, H. H., Nielsen, M. T. and Halkier, B. A. (2007). USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Res 35(7): e55.
- Hansen, N. B., Lübeck, M. and Lübeck, P. S. (2014). Advancing USER cloning into simple USER and nicking cloning. J Microbiol Methods 96: 42-49.
- Karimi, M., Depicker, A. and Hilson, P. (2007). Recombinational cloning with plant gateway vectors. Plant Physiol 145(4): 1144-1154.
- Marillonnet, S. and Grützner, R. (2020). Synthetic DNA Assembly Using Golden Gate Cloning and the Hierarchical Modular Cloning Pipeline. Curr Protoc in Mol Biol 130(1). https://doi.org/10.1002/cpmb.115.
- Patron, N. J., Orzaez, D., Marillonnet, S., Warzecha, H., Matthewman, C., Youles, M., Raitskin, O., Leveau, A., Farre, G., Rogers, C., et al. (2015). Standards for plant synthetic biology: a common syntax for exchange of DNA parts. New Phytol 208(1): 13-19.
- Vazquez-Vilar, M., Quijano-Rubio, A., Fernandez-del-Carmen, A., Sarrion-Perdigones, A., Ochoa-Fernandez, R., Ziarsolo, P., Blanca, J., Granell, A. and Orzaez, D. (2017). GB3.0: a platform for plant bio-design that connects functional DNA elements with associated biological data. Nucleic Acids Res 45(4): 2196-2209.
- Zhu, B., Cai, G., Hall, E. O. and Freeman, G. J. (2007). In-FusionTM assembly: Seamless engineering of multidomain fusion proteins, modular vectors, and mutations. Biotechniques 43(3): 354-359.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Pierdoná, F. G., Carbajal, Y., Vicente, M. H., Ferigolo, L. F. and Nogueira, F. T. S. (2022). Faster Bacterial Gene Cloning Using the Brick into the Gateway (BiG) Protocol. Bio-protocol 12(24): e4576. DOI: 10.21769/BioProtoc.4576.
Category
Molecular Biology > DNA > DNA cloning
Plant Science > Plant molecular biology > DNA > DNA modification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link