- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Di-ubiquitin Formation Assay and E3 Cooperation Assay
Published: Vol 12, Iss 21, Nov 5, 2022 DOI: 10.21769/BioProtoc.4547 Views: 1863
Reviewed by: Alexandros AlexandratosAndrew MacLeanAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Liposome Disruption Assay to Examine Lytic Properties of Biomolecules
John R. Jimah [...] Niraj H. Tolia
Aug 5, 2017 14266 Views
Activity-based Crosslinking to Identify Substrates of Thioredoxin-domain Proteins in Malaria Parasites
David W. Cobb [...] Vasant Muralidharan
Feb 20, 2022 2897 Views
Monitoring Protein Stability In Vivo Using an Intein-Based Biosensor
John S. Smetana [...] Christopher W. Lennon
Apr 20, 2025 1571 Views
Abstract
Ubiquitination is a post-translational modification conserved across eukaryotic species. It contributes to a variety of regulatory pathways, including proteasomal degradation, DNA repair, and cellular differentiation. The ubiquitination of substrate proteins typically requires three ubiquitination enzymes: a ubiquitin-activating E1, a ubiquitin-conjugating E2, and an E3 ubiquitin ligase. Cooperation between E2s and E3s is required for substrate ubiquitination, but some ubiquitin-conjugating E2s are also able to catalyze by themselves the formation of free di-ubiquitin, independently or in cooperation with a ubiquitin E2 variant. Here, we describe a method for assessing (i) di-ubiquitin formation by an E1 together with an E2 and an E2 variant, and (ii) the cooperation of an E3 with an E1 and E2 (with or without the E2 variant). Reaction products are assessed using western blotting with one of two antibodies: the first detects all ubiquitin conjugates, while the second specifically recognizes K63-linked ubiquitin. This allows unambiguous identification of ubiquitinated species and assessment of whether K63 linkages are present. We have developed these methods for studying ubiquitination proteins of Leishmania mexicana, specifically the activities of the E2, UBC2, and the ubiquitin E2 variant UEV1, but we anticipate the assays to be applicable to other ubiquitination systems with UBC2/UEV1 orthologues.
Keywords: UbiquitinBackground
Ubiquitination is a three-step process involving the sequential action of E1, E2, and E3 enzymes. In the first step, catalyzed by an E1 ubiquitin-activating enzyme, ubiquitin-adenylate is formed followed by ubiquitin transfer to a conserved cysteine on the enzyme. The second step involves a trans-thioesterification, in which the ubiquitin is transferred to a cysteine on the E2. In the third step, the E2, known as a ubiquitin-conjugating enzyme, cooperates with an E3 ubiquitin ligase to transfer ubiquitin to the target substrate, typically through the formation of an isopeptide bond to a lysine side chain (Zheng and Shabek, 2017). In many cases, target substrates can become poly-ubiquitinated through the attachment of further ubiquitin molecules. This attachment can take place on any of ubiquitin’s seven lysine side chains or on its amino terminal methionine, resulting in many permutations of ubiquitin conjugation (Zheng and Shabek, 2017). Specific ubiquitin linkages are associated with different cellular outcomes, although the understanding of this is incomplete (Komander and Rape, 2012). In an important “off-pathway” reaction, some E2s are also able to catalyze the formation of free di-ubiquitin, alone or with the assistance of a ubiquitin E2 variant (Wu et al., 2003; Wang et al., 2016; Burge et al., 2020).
The parasite Leishmania mexicana assumes different morphologies depending on its host: in the sand fly, it takes the motile promastigote form, whereas in a mammalian host, it takes the immotile amastigote form. Key to mammalian infection is the ability to undergo promastigote to amastigote differentiation. In L. mexicana, the E2 UBC2 and the ubiquitin E2 variant UEV1, which form a heterodimeric complex, are required for this process (Burge et al., 2020). Prior studies of the Saccharomyces cerevisiae UBC2 and UEV1 homologs UBC13 and MMS2, respectively, demonstrated that they could form K63-linked ubiquitin chains in the absence of an E3 (Hofmann and Pickart, 1999; McKenna et al., 2001; Wu et al., 2003; Andersen et al., 2005; Pastushok et al., 2005). By adapting methods used in these studies, the in vitro ubiquitin conjugation assays described here were established to delineate the ubiquitination activities of UBC2 and UEV1: firstly, to assess the cooperation of UBC2 and UEV1 in di-ubiquitin formation and, secondly, to assess the cooperation of UBC2 and UEV1 with E3 ubiquitin ligases (Figure 1). The method described here allows for the assessment of di-ubiquitin formation by an E2 and E2 variant pair and their collective cooperation with an E3 ligase. The assay uses western blotting specifically to detect ubiquitin conjugates. Blotting with an antibody specific to K63-conjugation also enables the assessment of whether the ubiquitin-conjugate products are linked at K63. Although we have not tested other antibodies, we anticipate that the method could easily be adapted to assay antibodies specific to other ubiquitin linkages.
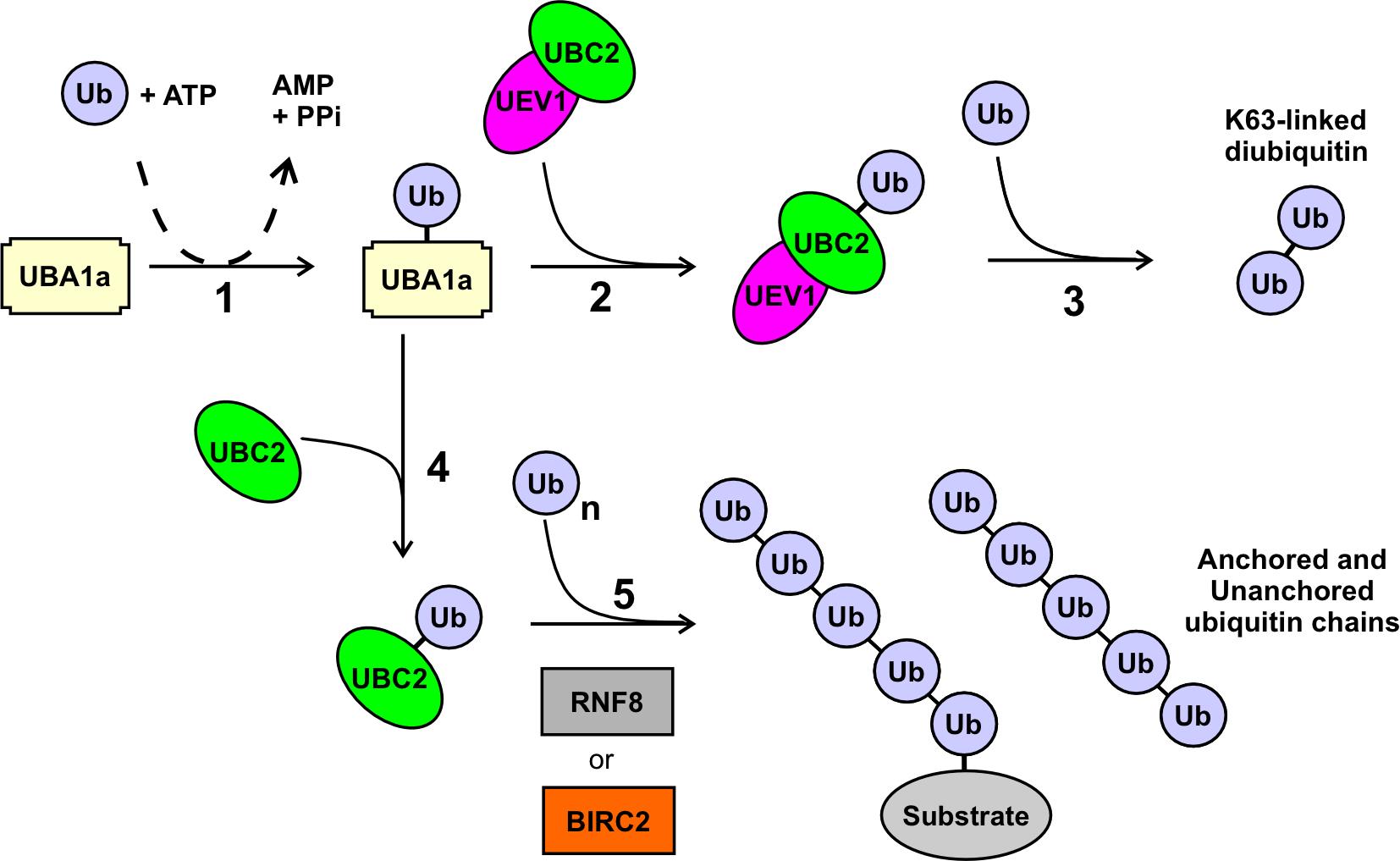
Figure 1. Ubiquitination outcomes mediated by UBC2 and UEV1. In the presence of ATP, ubiquitin (Ub) is activated and covalently attached in step 1 to the E1 UBA1a. In the presence of UEV1 and UBC2, the ubiquitin is transferred in step 2 to the UBC2 component of the UBC2–UEV1 heterodimer, and subsequently in step 3 onto a second ubiquitin molecule to form a K63-linked ubiquitin dimer, as described in Assay A. In the absence of UEV1, a UBC2–Ub complex is formed in step 4, which in the presence of an E3 such as RNF8 or BIRC2 forms polyubiquitin chains that may be unanchored or anchored to an unidentified substrate protein as described in Assay B. The linkage(s) in these chains are not known but are not K63-based.
Materials and Reagents
1.5 mL reaction tubes (Sarstedt, catalog number: 72.690.001)
100 mm square Petri dishes (Thermo Scientific, catalog number: 11349273)
HEPES (Fisher Bioreagents, catalog number: BP310-1)
NaCl (Fisher Chemicals, catalog number: S/3161/65)
MgCl2 (Sigma, catalog number: 63064)
DTT (Melford Laboratories, catalog number: D11000)
ATP (Sigma-Aldrich, catalog number: A2383)
Ubiquitin (R&D Systems, catalog number: U-100H-10M)
NuPAGE 4 to 12%, Bis-Tris, 1.0 mm, mini protein gels (Invitrogen, catalog number: NP0322BOX)
NuPAGE MES-SDS running buffer (20×) (Invitrogen, catalog number: NP0002)
NuPAGE LDS sample buffer (4×) (Invitrogen, catalog number: NP0007)
β-Mercaptoethanol (PanReac AppliChem, catalog number: A1108)
SeeBlue Plus2 pre-stained protein standard (Invitrogen, catalog number: LC5925)
PierceTM 20× TBS buffer (ThermoFisher, catalog number: 28358)
Tween-20 (Sigma, catalog number: P1379)
iBlot 2 nitrocellulose transfer stacks (Invitrogen, catalog number: IB23001)
Ethanol (VWR chemicals, catalog number: MFCD00003568)
Bovine serum albumin (BSA) (Sigma, catalog number: A3294)
Milk powder (Marvel)
Mono and polyubiquitylated conjugates, mAb (FK2) antibody (Ubiquigent, catalog number: 68-0121-500)
Ub-K63 mouse anti-human, clone: HWA4C4 (Affymetrix eBioscienceTM, catalog number: 14-6077-82)
HRP-conjugated anti-mouse antibody (Promega, catalog number: W4021)
Clarity Western ECL substrate (Bio-Rad, catalog number: 1705060)
Clarity Max Western ECL substrate (Bio-Rad, catalog number: 1705062)
Purified recombinant E1 (UBA1a, prepared under reducing conditions, storage: -70 °C) (Burge et al., 2020)
Purified recombinant E2 (UBC2, prepared under reducing conditions, storage: -70 °C) (Burge et al., 2020)
Purified recombinant E2 variant (UEV1, storage: -70 °C) (Burge et al., 2020)
Purified recombinant E3 (GST-BIRC2 or GST-RNF8, storage: -70 °C) (Ubiquigent, catalog number: 63-0015-025 or 63-0021-025)
di-ubiquitin, K63-linked (storage: -70 °C) (Ubiquigent, catalog number: 60-0107-010)
Reaction buffer (10×) (see Recipes)
SDS-PAGE sample buffer (4×) (see Recipes)
1× TBSt (see Recipes)
Equipment
Dry block heater (Grant QBD1)
Protein electrophoresis equipment (XCell SureLock Mini-Cell, EI0001)
Protein transfer system (iBlot 2 Dry Blotting System, Thermo Fisher Scientific)
Gel rocker
Microplate shaker (Grant, PMS-1000i)
Chemi-blot imager (ChemiDoc Imaging System, Bio-Rad)
Procedure
Di-ubiquitin formation assay
Prepare three reaction mixes in 1.5 mL tubes on ice containing the following reagents: 100 nM E1, 2.5 µM E2, 2.5 µM E2 variant, and 100 µM ubiquitin, in a buffer of 50 mM HEPES pH 7.5, 100 mM NaCl, 10 mM MgCl2, 2 mM DTT, and 5 mM ATP in a total volume of 40 µL. Typical reagent concentrations and volumes are given in Table 1. If a controlled reaction start is desired, the E1 or ATP can be omitted from the reaction mix during setup and added to initiate the reaction (mix by gently flicking the tube or stirring with a pipette tip during the final reagent addition). Three control reactions should also be performed, omitting either the E2, the E2 variant, or both the E2 and E2 variant, demonstrating that di-ubiquitin formation is not a consequence of activity from either the E2 or E2 variant alone, or from activity of the E1.
Table 1. Reaction composition
Component Working concentration Volume per reaction Final concentration E1 4 µM 1 µL 100 nM E2 25 µM 4 µL 2.5 µM E2 variant* 25 µM 4 µL 2.5 µM E3** 5 µM 8 µL 1.0 µM Ubiquitin 1167 µM 3.4 µL 100 µM Reaction buffer 500 mM HEPES pH 7.5 1 M NaCl
100 mM MgCl2
20 mM DTT
4 µL 50 mM HEPES pH 7.5
100 mM NaCl10 mM MgCl2
2 mM DTT
ATP 100 mM 2 µL 5 mM dH2O Up to 40 µL Up to 40 µL - *Can be excluded in the E3 cooperation assay
**Include only in the E3 cooperation assay
Incubate the reaction mixes in a dry block heater at 37 °C for 0 min, 30 min, and 90 min.
Stop the reactions by adding 1× SDS-PAGE sample buffer (13.3 µL) (see Recipes).
Heat the samples at 70 °C for 5 min using a dry block heater.
Run 20 µL of each sample on a NuPAGE mini protein gel in 1× NuPAGE MES-SDS running buffer using an electrophoresis system. Load 5 µL of SeeBlue Plus2 pre-stained protein standard to a single lane of the gel to enable molecular weight comparison of bands detected later during the western blot. Run the gel at 200 V until the lowest molecular weight marker is approximately 0.25 cm from the bottom of the gel (approximately 35 min). If analysis of both ubiquitin conjugates and K63-linked ubiquitin conjugates is desired, run two gels with 20 µL samples from each reaction on each gel. When assessing the presence of K63-linked ubiquitin, a sample of 100 ng K63-linked di-ubiquitin should be included on the gel, which will act as a positive control when blotting with the Ub-K63 antibody.
Incubate the gel in 20% ethanol in a square Petri dish for 10 min on a gel rocker at room temperature.
Transfer the proteins to a nitrocellulose membrane using the iBlot 2 Dry Blotting System and an iBlot 2 nitrocellulose transfer stack according to the manufacturer’s instructions. Perform the transfer at 20 V for 1 min, then 23 V for 4 min, and finally 25 V for 7 min.
Assessing the formation of ubiquitin conjugates using the FK2-antibody (Ubiquigent)
Block the membrane by covering it with 5% BSA dissolved in 1× TBSt (see Recipes) in a square Petri dish on a microplate shaker at 4 °C overnight.
Apply the mono and polyubiquitylated conjugates, mAb (FK2) antibody at a 1:1,000 dilution in 10 mL of 1% BSA in 1× TBSt in a square Petri dish. Incubate at 4 °C on a microplate shaker for 1 h.
Wash the membrane in 1× TBSt for 10 min three times at room temperature in a square Petri dish on a microplate shaker.
Apply the HRP-conjugated anti-mouse secondary antibody at 1:10,000 dilution in 10 mL of 5% BSA in 1× TBSt in a square Petri dish. Incubate at 4 °C on a microplate shaker for 1 h.
Wash the membrane in 1× TBSt for 10 min three times at room temperature in a square Petri dish on a microplate shaker operating at speed 4.
Apply clarity Western ECL substrate according to manufacturer’s instructions.
Measure chemiluminescence using a suitable imaging system (e.g., ChemiDoc imaging system, Bio-Rad).
Assessing the formation of K63-linked ubiquitin conjugates using the Ub-K63 antibody (ThermoFisher)
Block the membrane by covering it with 5% (w/v) milk dissolved in 1× TBSt. Incubate in a square Petri dish on a microplate shaker at 4 °C for 1 h.
Apply the Ub-K63 mouse antibody at 1:250 dilution in 5 ml of 5% milk in 1× TBSt in a square Petri dish. Incubate at 4 °C on a microplate shaker overnight.
Wash the membrane in 1× TBSt for 10 min three times at room temperature in a square Petri dish on a microplate shaker.
Apply the HRP-conjugated anti-mouse secondary antibody at 1:10,000 dilution in 10 mL of 5% (w/v) milk in 1× TBSt in a square Petri dish. Incubate at 4 °C on a microplate shaker overnight.
Wash the membrane in 1× TBSt for 10 min three times at room temperature in a square Petri dish on a microplate shaker.
Apply clarity Western ECL substrate according to manufacturer’s instructions.
Measure chemiluminescence using a suitable imaging system.
E3 cooperation assay
Prepare reaction mixes in 1.5 mL reaction tubes on ice containing the following reagents: 100 nM E1, 2.5 µM E2, 1 µM E3, and 100 µM ubiquitin in a buffer of 50 mM HEPES pH 7.5, 100 mM NaCl, 10 mM MgCl2, 2 mM DTT, and 5 mM ATP in a total volume of 40 µL. Where relevant, 2.5 µM of an E2 variant can also be included (e.g., when involved in di-ubiquitin formation in conjunction with an E2). Typical reagent concentrations and volumes are given in Table 1. If a controlled reaction start is desired, the E1 or ATP can be omitted from the reaction mix during setup and added to initiate the reaction (mix by gently flicking the tube or stirring with a pipette tip during the final reagent addition).
To determine whether the observed activity is due to the E3, four control reactions should be carried out, omitting either the E3, the E2, the E2 variant, or both the E2 and E2 variant. This allows for the assessment of any activity that may be a consequence of the E1 and E2s in the absence of the E3, and also whether the E3 works in cooperation with the E2 or E2 variant, alone or in combination.
Incubate reactions in a dry block heater at 30 °C for 60 min or other times as required.
Carry out steps A3–A21.
Data analysis
The FK2 antibody detects all ubiquitin conjugates, which are observed as chemiluminescent bands on the western blot. Comparison of these bands with the pre-stained ladder allows confirmation of di-ubiquitin formation. This is seen in Figure 2A, where incubation of Leishmania UBC2 and UEV1 for 30 or 90 min resulted in the formation of a clear band for di-ubiquitin (minor bands were also observed for UBA1a-Ub and UBC2-Ub complexes because these also contain a conjugated ubiquitin). The FK2 antibody can also be used to detect substrate and/or poly-ubiquitination, as desired in the E3 cooperation assay (Figure 3A). In this example, multiple ubiquitin conjugate bands were seen when Leishmania UBC2 was incubated with the human E3s BIRC2 or RNF8.
Blotting with the Ub-K63 antibody allows the specific assessment of the formation of K63-linked ubiquitin conjugates (Figure 2B and Figure 3B). Thus, assessment of the same reaction products using both the FK2 and Ub-K63 antibodies enables the analysis of whether conjugates detected by the FK2 antibody are K63-linked. For example, in Figure 2, the di-ubiquitin band that was observed when blotting with the FK2 antibody (Figure 2A) was also observed when blotting with the Ub-K63 antibody (Figure 2B), indicating that the di-ubiquitin formed by UBC2 and UEV1 is K63-linked. However, in the E3 cooperation assay (Figure 3), conjugates produced by incubation of UBC2 with BIRC2 or RNF8 that were detected by the FK2 antibody (Figure 3A) were not detected by the Ub-K63 antibody (Figure 3B), suggesting that these conjugates are not K63-linked.
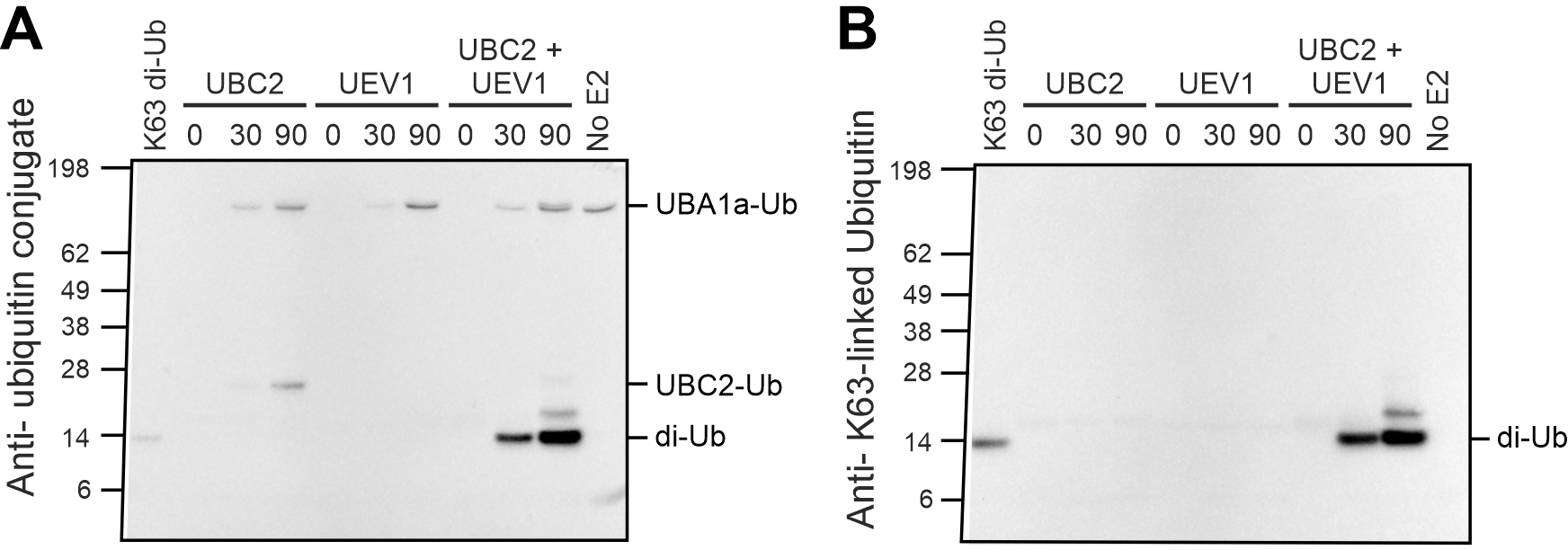
Figure 2. Di-ubiquitin formation assay. A) Blot using FK2 ubiquitin conjugate antibody. B) Blot using K63-linked ubiquitin antibody. Reproduced from Burge et al. (2020).
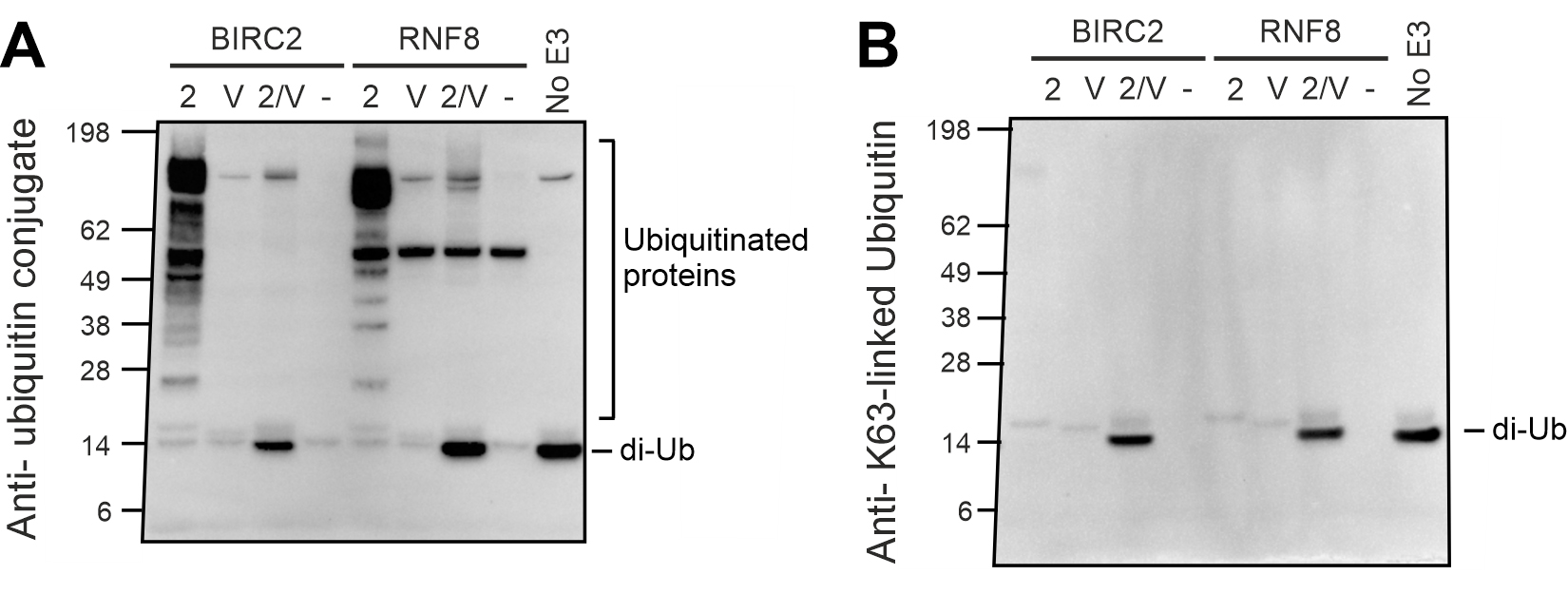
Figure 3. E3 Cooperation assay. A) Blot using FK2 ubiquitin conjugate antibody. B) Blot using K63-linked ubiquitin antibody. Reproduced from Burge et al. (2020).
Notes
UBA1a and UBC2 need to be purified under reducing conditions to maintain catalytic activity: 5 mM β-mercaptoethanol was included in the purification buffers for nickel affinity purification and 2 mM DTT or 1 mM TCEP for size exclusion chromatography. Proteins were stored at -70 °C in buffers containing 2 mM DTT or 1 mM TCEP (Note: UBA1a and UBC2 purified in buffers without reducing agents were not catalytically active). Avoid freeze-thaw cycles to increase the storage lifetime of the proteins.
Successful assays were achieved with the following changes in gel-running and transfer conditions: firstly, running samples on 4%–20% Mini-Protean TGX gels (Bio-Rad) in a running buffer of 25 mM Tris and 192 mM glycine pH 8.3, at 200 V for 35 min; secondly, incubating the gel, a nitrocellulose membrane, and filter paper in a transfer buffer of 25 mM Tris, 192 mM glycine, and 10% v/v methanol for 30 min before transfer using a Trans-Blot SD semi-dry transfer cell (Bio-Rad) at 25 V for 25 min.
If low activity is seen in the E3 cooperation assay, try increasing the incubation temperature to 37 °C.
Recipes
Reaction buffer (10×)
500 mM HEPES pH 7.5
1 M NaCl
100 mM MgCl2
20 mM DTT
SDS-PAGE sample buffer (4×)
95% NuPAGE LDS sample buffer (4×)
5% β-mercaptoethanol
1× TBSt
5% (v/v) PierceTM 20× TBS buffer
0.05% (v/v) Tween-20
94.95% (v/v) deionized H2O
Acknowledgments
This protocol was adapted from the research published in Burge et al. (2020). We would like to thank Ubiquigent for providing a 3 month internship for RB, during which this protocol was developed. This work was supported by a Medical Research Council Studentship to RB (MRC MR/N018230/1) and the Wellcome Trust to JCM (200807/Z/16/Z).
Competing interests
The authors declare no competing interests
References
- Andersen, P. L., Zhou, H., Pastushok, L., Moraes, T., McKenna, S., Ziola, B., Ellison, M. J., Dixit, V. M. and Xiao, W. (2005). Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A. J Cell Biol 170(5): 745-755.
- Burge, R. J., Damianou, A., Wilkinson, A. J., Rodenko, B. and Mottram, J. C. (2020). Leishmania differentiation requires ubiquitin conjugation mediated by a UBC2-UEV1 E2 complex. PLoS Pathog 16(10): e1008784.
- Hofmann, R. M. and Pickart, C. M. (1999). Non-canonical MMS2-Encoded Ubiquitin-Conjugating Enzyme Functions in Assembly of Novel Polyubiquitin Chains for DNA Repair. Cell 96: 645-53.
- Komander, D. and Rape, M. (2012). The ubiquitin code. Annu Rev Biochem 81: 203-229.
- McKenna, S., Spyracopoulos, L., Moraes, T., Pastushok, L., Ptak, C., Xiao, W. and Ellison, M. J. (2001). Noncovalent interaction between ubiquitin and the human DNA repair protein Mms2 is required for Ubc13-mediated polyubiquitination. J Biol Chem 276(43): 40120-6.
- Pastushok, L., Moraes, T. F., Ellison, M. J. and Xiao, W. (2005). A single Mms2 "key" residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys63 ubiquitin conjugation complex. J Biol Chem 280(18): 17891-17900.
- Wang, S., Cao, L. and Wang, H. (2016). Arabidopsis ubiquitin-conjugating enzyme UBC22 is required for female gametophyte development and likely involved in Lys11-linked ubiquitination. J Exp Bot 67(11): 3277-3288.
- Wu, P. Y., Hanlon, M., Eddins, M., Tsui, C., Rogers, R. S., Jensen, J. P., Matunis, M. J., Weissman, A. M., Wolberger, C. and Pickart, C. M. (2003). A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J 22(19): 5241-5250.
- Zheng, N. and Shabek, N. (2017). Ubiquitin Ligases: Structure, Function, and Regulation. Annu Rev Biochem 86: 129-157.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Burge, R. J., Jameson, K. H., Wilkinson, A. J. and Mottram, J. C. (2022). In vitro Di-ubiquitin Formation Assay and E3 Cooperation Assay. Bio-protocol 12(21): e4547. DOI: 10.21769/BioProtoc.4547.
Category
Microbiology > Microbial biochemistry > Protein > Activity
Biochemistry > Protein > Activity
Biological Sciences > Biological techniques > Microbiology techniques
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link