- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
X-ray Crystallography: Seeding Technique with Cytochrome P450 Reductase
Published: Vol 12, Iss 21, Nov 5, 2022 DOI: 10.21769/BioProtoc.4546 Views: 2306
Reviewed by: Joana Alexandra Costa ReisLaura AlviginiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Localization and Topology of Thylakoid Membrane Proteins in Land Plants
Salar Torabi [...] Jörg Meurer
Dec 20, 2014 13393 Views

Heterologous Expression and Purification of Catalytic Domain of CESA1 from Arabidopsis thaliana
Venu Gopal Vandavasi and Hugh O’ Neill
Oct 20, 2016 7529 Views

Expression, Purification and Crystallization of Recombinant Arabidopsis Monogalactosyldiacylglycerol Synthase (MGD1)
Joana Rocha [...] Christelle Breton
Dec 20, 2016 8824 Views
Abstract
Cytochrome P450 reductase (CPR) is a multi-domain protein that acts as a redox partner of cytochrome P450s. The CPR contains a flavin adenine dinucleotide (FAD)–binding domain, a flavin mononucleotide (FMN)-binding domain, and a connecting domain. To achieve catalytic events, the FMN-binding domain needs to move relative to the FAD-binding domain, and this high flexibility complicates structural determination in high-resolution by X-ray crystallography. Here, we demonstrate a seeding technique of sorghum CPR crystals for resolution improvement, which can be applied to other poorly diffracting protein crystals. Protein expression is completed using an E. coli cell line with a high protein yield and purified using chromatography techniques. Crystals are screened using an automated 96-well plating robot. Poorly diffracting crystals are originally grown using a hanging drop method from successful trials observed in sitting drops. A macro seeding technique is applied by transferring crystal clusters to fresh conditions without nucleation to increase crystal size. Prior to diffraction, a dehydration technique is applied by serial transfer to higher precipitant concentrations. Thus, an increase in resolution by 7 Å is achieved by limiting the inopportune effects of the flexibility inherent to the domains of CPR, and secondary structures of SbCPR2c are observed.
Graphical abstract:
Background
Cytochrome P450 reductase (CPR, EC 1.6.2.4) is located at the cytosol side of the endoplasmic reticulum membrane in eukaryotic cells, acting as the electron donor to cytochrome P450s and other heme proteins (Phillips and Langdon, 1962). It has been known that there are one to three CPR genes in most vascular plants (Jensen and Møller, 2010). The CPR contains a flavin adenine dinucleotide (FAD)/NADPH-binding domain, a flavin mononucleotide (FMN)-binding domain, and a connecting (or linker) domain with a flexible hinge region that links these two domains (Munro et al., 2001; Wang et al., 1997). We have reported on the structural flexibility of sorghum CPR isoforms (Zhang et al., 2022b) and the rapid interdomain opening and closing motion that resulted in fast interdomain electron transfer in SbCPR (Zhang et al., 2022a). Especially in the SbCPR2c isoform, which was crystallized in full-length, the FMN-binding domain was not resolved due to the aforementioned flexibility. Such flexibility causes uncertainty of the residue atomic coordinates and dihedral angles of the protein crystal, which leads to local disordered structure and overall low-resolution diffraction.
Crystallization of diffraction quality crystals is the largest bottleneck in protein structure determination (Holcomb et al., 2017). These difficulties have been overcome through the use of various techniques. Seeding uses previously nucleated crystals to initiate the growth of larger crystals in a fresh drop where protein concentration has not yet been depleted by the many nucleation sites of the small crystals (Rhodes, 2006). Micro streaking operates by running a fiber over an existing crystal—often a whisker from an animal—through a fresh drop to increase growth of the nucleated particles in a supersaturated drop (Stura and Wilson, 1991). In contrast, a macro seeding technique utilizes larger crystals, and thus requires more precision as a chosen crystal is washed and transferred from its original drop to a new mother liquor. This method tends to be more meticulous and can fracture the original crystal, causing smaller fragments to be placed into the new drop, which induces micro seeding (Zhu et al., 2005). Seeding techniques have allowed for growth of larger crystals and increased resolution of diffraction; cellobiohydrolase II (CBHII) crystallization was first achieved by using seeding to avoid formation of microcrystals and diffraction greater than 2.0 Å (Bergfors et al., 1989). High mosaicity and cracking/sliding of the lattice caused by impurities have been overcome by using seeding to reduce crystal deficits and improve the resolution of crystals (Caylor et al., 1999). Another method for improving crystal lattice packing is the dehydration of the crystalline to reduce water contents and to consequently confer tighter packing (Timasheff, 1995). Dehydration can be accomplished via exposure to the atmosphere or via serial dilution into higher cryoprotectant-containing solutions (Heras and Martin, 2005). Crystal dehydration has been one of the most successful post crystallization treatments in increasing resolution. Notably, the resolution of disulfide bond isomerase was improved from 7 to 2.6 Å and from 12 to 2.6 Å in E. coli YbgL (Abergel, 2004; Haebel et al., 2001).
We incorporated a macromolecule seeding technique—additive solution optimization—together with dehydration to improve the resolution of the SbCPR2c crystal structure from approximately 12 to 4.5 Å. This enabled the determination of the secondary structure for SbCPR2c. It is worth noting, however, that crystal growth is a very tedious process with repeatability differing in trials due to miniscule changes of parameters and the complexity of the system. Despite the aforementioned difficulties, our protocol of seeding and dehydration could be widely applied to resolve the resolution problems to a variety of crystals hindered in resolution by their dynamic disorder.
Materials and Reagents
MRC 2-well crystallization plate (Swissci, Hampton Research, catalog number: HR3-082)
24-well XRL plate (Molecular Dimensions, catalog number: MD3-11)
Additive screen (Hampton Research, catalog number: HR2-428)
Microscope cover glass (Fisher Scientific, Fisherbrand, catalog number: 19803)
Glass capillary (Charles Super Company, catalog number: 02-SG)
Amicon 8050 ultrafiltration cell (Amicon, catalog number: UFSC05001)
30 kDa ultrafiltration discs (EMD Millipore Corporation, Biomax, catalog number: PBTK04310)
Escherichia coli Rosetta 2 (DE3) competent cells (Sigma-Aldrich, Millipore Sigma, catalog number: 71402)
pET-30a(+) vector (Sigma-Aldrich, Millipore Sigma, catalog number: 69909)
Sorghum bicolor CPR2c gene (Sobic.006G245400) (synthesized by Genscript)
Kanamycin (IBI Scientific, catalog number: IB02120)
IPTG (GoldBio, catalog number:I2481C100)
NADP+ (Alfa Aesar, catalog number: T23B015)
PEG 3350 (Sigma-Aldrich, catalog number: P4338)
Nickel-NTA resin (G-biosciences, catalog number: 786-940)
Index crystal screen kit (Hampton Research, catalog number: HR2-144)
Luria-Bertani (LB) medium (see Recipes)
Lysis/wash buffer (see Recipes)
Elution buffer (see Recipes)
Hydroxyapatite column buffer A (see Recipes)
Hydroxyapatite column buffer B (see Recipes)
Final buffer (see Recipes)
Crystallization buffer (crystal screen F12) (see Recipes)
Equipment
Crystal Phoenix (Art Robbins Instruments)
ÄKTA pure (GE Healthcare, ÄKTA pure, catalog number: 29014834)
Model 450 sonicator (Branson Ultrasonics, catalog number: 22-309783)
Software
HKL2000 (https://hkl-xray.com/hkl-2000)
Phenix (https://phenix-online.org/)
Coot (https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/)
Procedure
Gene cloning
Clone the Sorghum bicolor CPR2c gene (Sobic.006G245400) with a N-terminal 6×His tag and without the native N-terminal hydrophobic membrane–anchoring region (Δ1–50) into vector pET-30a(+) for overexpression.
Cell transformation
Put E. coli Rosetta 2 (DE3) competent cells on ice.
Add 1 μL of plasmid (20 ng/μL) into 20 μL of competent cells.
Incubate on ice for 5 min.
Heat shock for 30 s.
Incubate on ice for 2 min.
Add 80 μL of LB medium to competent cells.
Shake at 37 °C at 220 rpm for 1 h.
Plate all the cells on an LB agar plate with 50 ng/μL of kanamycin and incubate overnight.
Protein expression
Pick a single colony from the LB agar plate and grow in 5 mL of LB medium supplemented with 50 ng/μL kanamycin overnight at 37 °C.
Grow 20 mL of starting culture from the 5 mL culture with 50 ng/μL kanamycin overnight at 37 °C.
Transfer 20 mL of starting culture to 3 L of LB medium with 50 ng/μL kanamycin.
Shake at 220 rpm and 37 °C until reaching a OD600 of 0.4–0.6.
Cool down the medium to 25 °C and add 0.5 mM of IPTG to induce.
Induce the cells at 25 °C for 16 h.
Harvest the cells at 8,000 × g and 4 °C for 10 min.
Protein purification
Resuspend the cells with lysis buffer.
Set the output control at 9 and duty cycle at 90%. Sonicate the cells for 30 s followed by resting on ice for 5 min until homogenous.
Centrifuge at 34,000 × g and 4 °C for 1 h to remove the cell debris.
Load the clear lysate on a gravity column containing 20 mL of nickel-NTA resin pre-equilibrated with lysis buffer.
Wash with 200 mL of the same buffer.
Elute the protein with 100 mL of elution buffer.
Concentrate and buffer exchange against hydroxyapatite column buffer A using an Amicon 8050 ultrafiltration cell with a 30 kDa cutoff membrane.
Load the buffer-exchanged protein on the hydroxyapatite column.
Wash with 100 mL of 5% hydroxyapatite column buffer B.
Elute protein with 10% hydroxyapatite column buffer B.
Concentrate and buffer exchange against final buffer using an Amicon 8050 ultrafiltration cell with a 30 kDa cutoff membrane.
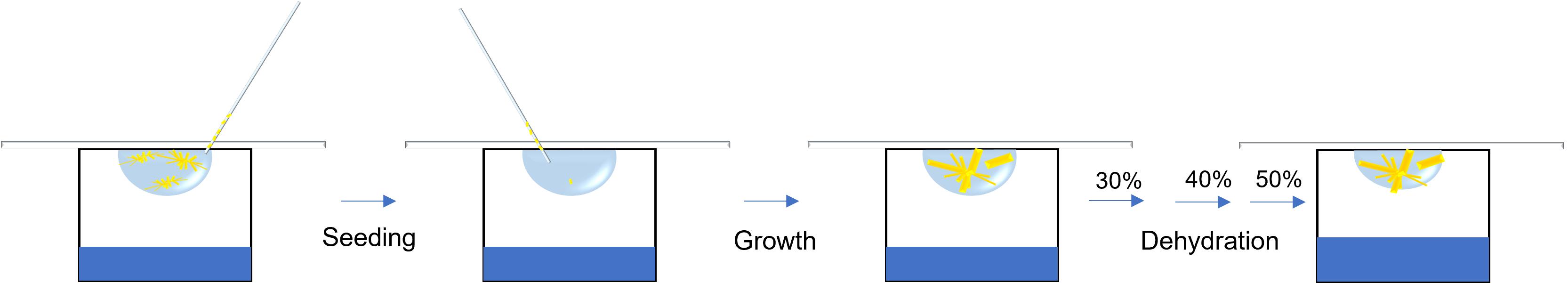
Initial crystallization setup
Concentrate the purified protein to 30 mg/mL, determined by the molar extinction coefficient of CPR (21.4 mm−1 cm−1).
Add 1.2 mM of NADP+ prior to crystallization.
Run Crystal Phoenix with an MRC 2-well crystallization plate with the index crystal screen kit and set up at room temperature (22 °C). The drop contains 0.2 μL of protein solution and 0.2 μL of screening solution.
Small crystals show up in condition F12 (see crystallization buffer in Recipes) in two days (Figure 1A).
Reproduce and optimize the crystal by mixing 1.6 μL of crystallization buffer, 2 μL of protein solution, and 0.4 μL of additive screen on a cover glass.
Set up the hanging drop vapor diffusion in 24-well plates with 0.5 mL of crystallization buffer in reservoir.
Needle-shaped crystals show up with additive screen condition 31 (1,5-diaminopentane dihydrochloride) in 3–5 days (Figure 1B).
Crystal seeding
Mix 2.4 μL of crystallization buffer, 3 μL of protein solution, and 0.6 μL of additive screen.
Suck one needle crystal from the previous drop, place into the new 6 μL crystallization drop, and set up the hanging drop.
Large crystals show up in one week (Figure 1C).
Transfer the cover glass to the reservoir filled with 0.5 mL of 30% (v/w), 40% (v/w), and 50% (v/w) PEG 3350 every third day for sequential dehydration.
Fast freeze the dehydrated crystals in liquid nitrogen for X-ray diffraction after equilibrating in 50% (v/w) PEG 3350 for two days.
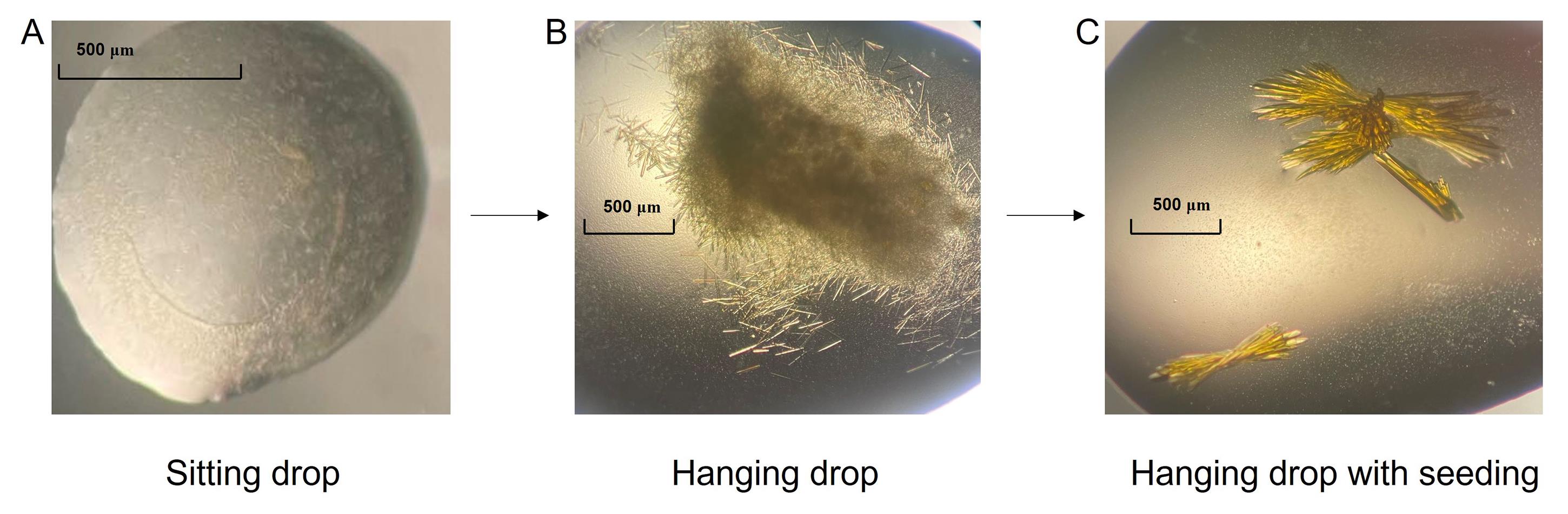
Figure 1. Optimization of SbCPR2c crystals. (A) Crystals obtained from the hit [index condition F12 (see crystallization buffer in Recipes)] of screening. (B) Crystals obtained from hanging drop with additive solution optimization. (C) Crystals obtained from seeding technique.Data collection and structure determination
Diffract the crystals at the beamline by using beam size 100 × 100 μm, 10 s, and 1 degree per frame and 180 degrees in total.
Index, integrate, and scale the data using HKL2000.
Do molecular replacement with the proper model [in our case, SbCPR2b (PDBID: 7SUX)] with PHENIX, run AutoBuild, and refine the structure with PHENIX and Coot.
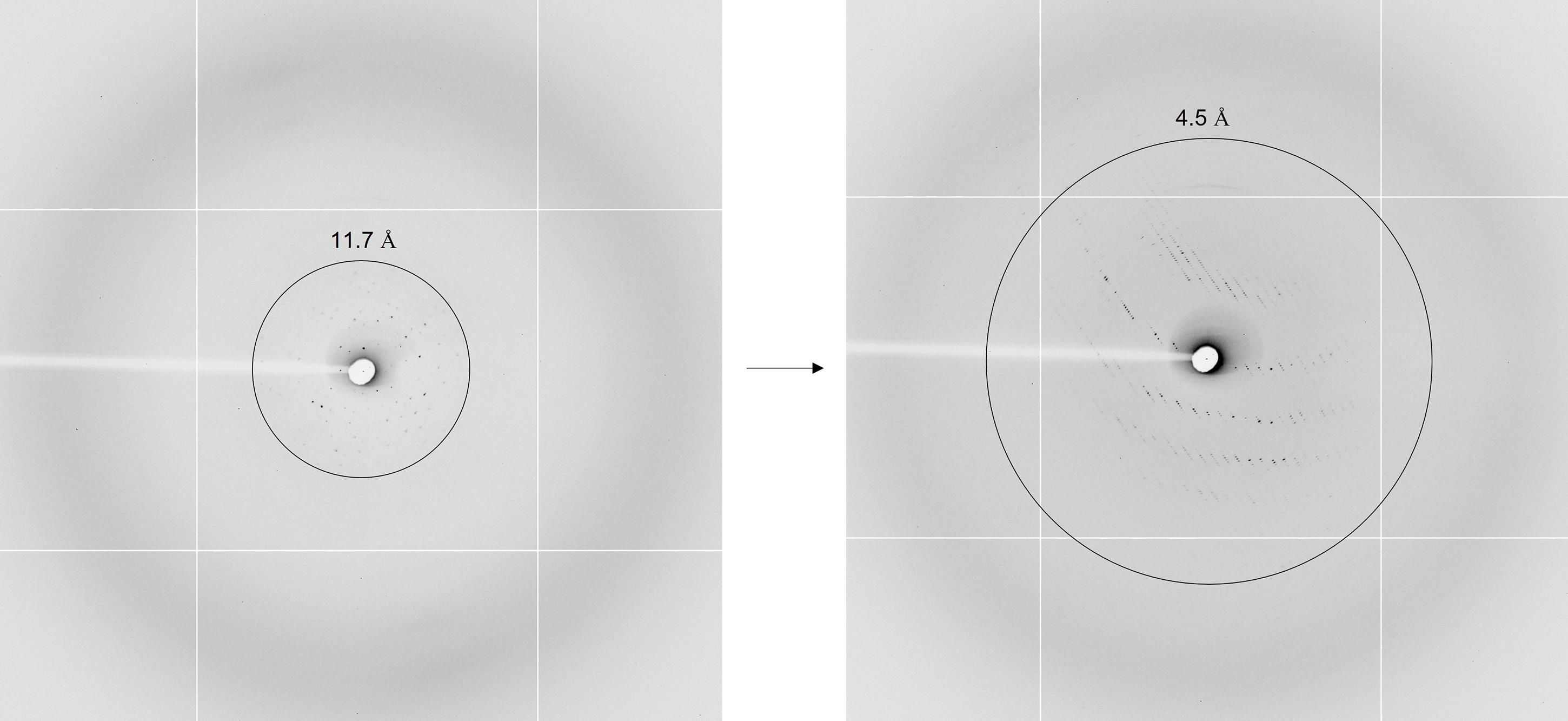
Figure 2. The resolution of the crystal has been improved from 11.7 Å (from non-seeding hanging drop crystals) to 4.5 Å (seeding crystals)
Data analysis
The diffraction data was indexed, integrated, and scaled with HKL2000. The molecular replacement was performed with the SbCPR2b structure (PDBID: 7SUX) and with Phaser-MR (full-featured) in the PHENIX software. The number of molecules in an asymmetric unit was calculated with Matthews coefficients (Matthews, 1975). AutoBuild was then run for model building with the SbCPR2c sequence. A rigid body refinement was completed before full refinement of XYZ (reciprocal-space), XYZ (real-space), and individual B-factors with phenix.refine. The individual residues were refined in Coot.
Notes
The crystallization and additive solutions are specific for SbCPR2c. It is necessary to initially screen for the optimal condition for the protein of interest. If crystals are of poor shape, using additive screen solution can rectify poor crystal edges.
Using additive screen solution may cause precipitation. When placing the crystal seed, you should avoid disruption to the drop, which can mix the solutions and will impede crystal growth.
Recipes
Luria-Bertani (LB) medium
Reagent Final concentration Amount Tryptone 10 g/L 10 g Yeast extract 5 g/L 5 g NaCl 10 g/L 10 g dH2O n/a n/a Total n/a 1,000 mL Lysis/wash buffer
Reagent Final concentration Amount NaCl 300 mM 17.532 g Tris 50 mM 6.057 g Imidazole 20 mM 1.36 g dH2O n/a n/a HCl (12 M) n/a As needed to adjust pH to 8 Total n/a 1,000 mL Elution buffer
Reagent Final concentration Amount NaCl 300 mM 17.532 g Tris 50 mM 6.057 g Imidazole 250 mM 17.02 g dH2O n/a n/a HCl (12 M) n/a As needed to adjust pH to 8 Total n/a 1,000 mL Hydroxyapatite column buffer A
Reagent Final concentration Amount KH2PO4 5 mM 0.68 g NaOH n/a As needed to adjust pH to 6.8 dH2O n/a n/a Total n/a 1,000 mL Hydroxyapatite column buffer B
Reagent Final concentration Amount KH2PO4 500 mM 68 g NaOH n/a As needed to adjust pH to 6.8 dH2O n/a n/a Total n/a 1,000 ml Final buffer
Reagent Final concentration Amount Hydroxyapatite column buffer B 50 mM 100 mL dH2O n/a 900 mL Total n/a 1,000 mL Crystallization buffer (crystal screen F12)
Reagent Final concentration Amount NaCl (2 M) 200 mM 5 mL HEPES (1 M, pH 7.5) 100 mM 5 mL dH2O n/a n/a PEG 3350 (50%, m/v) 25% (w/v) 25 mL Total n/a 50 mL
Acknowledgments
Original research was supported by grant from NSF (CHE-1804699, MCB-2043248) and Murdock Charitable Trust (CHK). This research used resources of the Advanced Light Source (beamline 5.0.2 and beamline 5.0.3), which is a DOE Office of Science User Facility under contract no. DE-AC02-05CH11231. The protocol was derived from original research paper (Zhang et al., 2022b).
Competing interests
There is no competing interest.
References
- Abergel, C. (2004). Spectacular improvement of X-ray diffraction through fast desiccation of protein crystals. Acta Crystallogr D Biol Crystallogr 60(Pt 8): 1413-1416.
- Bergfors, T., Rouvinen, J., Lehtovaara, P., Caldentey, X., Tomme, P., Claeyssens, M., Pettersson, G., Teeri, T., Knowles, J. and Jones, T. A. (1989). Crystallization of the core protein of cellobiohydrolase II from Trichoderma reesei. J Mol Biol 209(1): 167-169.
- Caylor, C. L., Dobrianov, I., Lemay, S. G., Kimmer, C., Kriminski, S., Finkelstein, K. D., Zipfel, W., Webb, W. W., Thomas, B. R., Chernov, A. A., et al. (1999). Macromolecular impurities and disorder in protein crystals. Proteins 36(3): 270-281.
- Jensen, K. and Møller, B. L. (2010). Plant NADPH-cytochrome P450 oxidoreductases. Phytochemistry 71: 132-141.
- Haebel, P. W., Wichman, S., Goldstone, D. and Metcalf, P. (2001). Crystallization and initial crystallographic analysis of the disulfide bond isomerase DsbC in complex with the alpha domain of the electron transporter DsbD. J Struct Biol 136(2): 162-166.
- Heras, B. and Martin, J. L. (2005). Post-crystallization treatments for improving diffraction quality of protein crystals. Acta Crystallogr D Biol Crystallogr 61(Pt 9): 1173-1180.
- Holcomb, J., Spellmon, N., Zhang, Y., Doughan, M., Li, C. and Yang, Z. (2017). Protein crystallization: Eluding the bottleneck of X-ray crystallography. AIMS Biophys 4(4): 557-575.
- Matthews, B. W. (1975). Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta 405(2): 442-451.
- Munro, A. W., Noble, M. A., Robledo, L., Daff, S. N. and Chapman, S. K. (2001). Determination of the redox properties of human NADPH-cytochrome P450 reductase. Biochemistry 40(7): 1956-1963.
- Phillips, A. H. and Langdon, R. G. (1962). Hepatic triphosphopyridine nucleotide-cytochrome c reductase: isolation, characterization, and kinetic studies. J Biol Chem 237: 2652-2660.
- Rhodes, G. (2006). Chapter 3 - Protein Crystals. In: Rhodes, G. (Ed.) Crystallography Made Crystal Clear (Third Edition). Academic Press, 31-47.
- Stura, E. A. and Wilson, I. A. (1991). Applications of the streak seeding technique in protein crystallization. J Cryst Growth 110(1): 270-282.
- Timasheff, S. N. (1995). Solvent stabilization of protein structure. Methods Mol Biol 40: 253-269.
- Wang, M., Roberts, D. L., Paschke, R., Shea, T. M., Masters, B. S. S. and Kim, J.-J. P. (1997). Three-dimensional structure of NADPH–cytochrome P450 reductase: prototype for FMN-and FAD-containing enzymes. Proc Natl Acad Scie U S A 94(16): 8411-8416.
- Zhang, B., Kang, C. and Davydov, D. R. (2022a). Conformational Rearrangements in the Redox Cycling of NADPH-Cytochrome P450 Reductase from Sorghum bicolor Explored with FRET and Pressure-Perturbation Spectroscopy. Biology (Basel) 11(4).
- Zhang, B., Munske, G. R., Timokhin, V. I., Ralph, J., Davydov, D. R., Vermerris, W., Sattler, S. E. and Kang, C. (2022b). Functional and structural insight into the flexibility of cytochrome P450 reductases from Sorghum bicolor and its implications for lignin composition. J Biol Chem 298(4): 101761.
- Zhu, D. Y., Zhu, Y. Q., Xiang, Y. and Wang, D. C. (2005). Optimizing protein crystal growth through dynamic seeding. Acta Crystallogr D Biol Crystallogr 61(Pt 6): 772-775.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Zhang, B., Lewis, J. A., Hazra, R. and Kang, C. (2022). X-ray Crystallography: Seeding Technique with Cytochrome P450 Reductase. Bio-protocol 12(21): e4546. DOI: 10.21769/BioProtoc.4546.
- Zhang, B., Munske, G. R., Timokhin, V. I., Ralph, J., Davydov, D. R., Vermerris, W., Sattler, S. E. and Kang, C. (2022b). Functional and structural insight into the flexibility of cytochrome P450 reductases from Sorghum bicolor and its implications for lignin composition. J Biol Chem 298(4): 101761.
Category
Plant Science > Plant biochemistry > Protein > Structure
Biophysics > X-ray crystallography
Biochemistry > Protein > Structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









