- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Carbamoyltransferase Enzyme Assay: In vitro Modification of 5-hydroxymethylcytosine (5hmC) to 5-carbamoyloxymethylcytosine (5cmC)
Published: Vol 12, Iss 17, Sep 5, 2022 DOI: 10.21769/BioProtoc.4496 Views: 1982
Reviewed by: Gal HaimovichQin TangAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
A Novel Protein Purification Approach Using Elastin-Like Polypeptides (ELP) With His-Tag Assistance
Young Kee Chae and Han Bin Shin
Jun 20, 2025 3088 Views
Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2159 Views
Prokaryotic Expression and Purification of the hSox2-HMG Domain
Lijie Yang [...] Jingjun Hong
Aug 20, 2025 2362 Views
Abstract
Nucleic acids in living organisms are more complex than the simple combinations of the four canonical nucleotides. Recent advances in biomedical research have led to the discovery of numerous naturally occurring nucleotide modifications and enzymes responsible for the synthesis of such modifications. In turn, these enzymes can be leveraged towards toolkits for DNA and RNA manipulation for epigenetic sequencing or other biotechnological applications. Here, we present the protocol to obtain purified 5-hydroxymethylcytosine carbamoyltransferase enzymes and the associated assays to convert 5-hydroxymethylcytosine to 5-carbamoyloxymethylcytosine in vitro. We include detailed assays using DNA, RNA, and single nucleotide/deoxynucleotide as substrates. These assays can be combined with downstream applications for genetic/epigenetic regulatory mechanism studies and next-generation sequencing purposes.
Keywords: DNA modificationBackground
Modification and hyper modification of canonical nucleotides can be found in all domains of life. In bacteriophages, nucleotide modifications are often used as a strategy to evade bacterial immunity, most notably the restriction/modification system. Thus, bacteriophages tend to have a broad array of modifications present throughout their genomes. For example, the genome of T4 bacteriophages uses hydroxymethylated deoxycytidine triphosphate (dCTP) as substrate during DNA synthesis, effectively replacing dC with 5dhmC. Incorporated 5dhmC is further post-synthetically glycosylated, resulting in DNA containing glycosylated 5-hydroxymethylcytosine (Lehman and Pratt, 1960). Recent studies have aimed at expanding the repertoire of known modifications by mining the microbiomes for DNA-modifying enzymes (Ferrer et al., 2005).
In a previous study, we developed the Metagenomics Genome–Phenome Association (MetaGPA) framework to associate genetic data with phenotypic traits at the level of an entire microbiome (Yang et al., 2021). As a case study, we applied MetaGPA to sewage and ocean microbiome samples and successfully identified enzymes associated with cytosine modifications in the DNA. Experimental follow-up on these associations allowed us to identify and validate a novel DNA/RNA modifying enzyme, the 5-hydroxymethylcytosine carbamoyltransferase. This enzyme catalyzes the formation of a previously unknown form of cytosine modification, the 5-carbamoyloxymethylcytosine, using 5-hydroxymethylcytosine in DNA, RNA, or single nucleotides as substrates. Here, we describe the protocol for cloning, expressing, and purifying the enzyme from E. coli. We also describe the protocol for assaying this enzyme in vitro on both nucleic acid polymers and single nucleotides, which greatly expands its potential applications. As shown previously (Yang et al., 2021), the carbamoyltransferase reaction can be included in a next-generation sequencing-based method for genomic mapping of modified cytosines. Other possible applications include genetic engineering, DNA/RNA polymerase kinetics, and post-synthetic modification regulations.
Materials and Reagents
Consumables
Low retention 10 μL tips (VWR, catalog number: 89135-572)
Low retention 200 μL tips (VWR, catalog number: 76175-406)
Low retention 1,000 μL tips (VWR, catalog number: 76175-412)
1.5 mL microcentrifuge tube (Eppendorf, catalog number: 022363204)
8 strip 0.2 mL PCR tubes (VWR, catalog number: 20170-010)
15 mL centrifuge tubes (VWR, catalog number: 21008-103)
50 mL centrifuge tubes (VWR, catalog number: 21008-242)
5 mL serological pipets (VWR, catalog number: 89130-908)
10 mL serological pipets (VWR, catalog number:89130-888)
25 mL serological pipets (VWR, catalog number: 89130-900)
Weigh boats (any source)
Gloves (any source)
Oligo clean & concentrator kit (Zymo Research, catalog number: D4060)
Oligo clean-up and concentrator kit (Norgen Biotek, catalog number: 34100)
For carbamoyltransferase enzyme expression and purification:
Bacteria culture tube (any source)
Carbamoyltransferase plasmid pET28-Kan-his-CT (available upon request)
Competent DH5-alpha E. coli cells (New England Biolabs, catalog number: C2987H)
T7 Express competent E. coli cells (New England Biolabs, catalog number: C2566)
2 L flask (VWR)
Kanamycin stock solution, 40 mg/mL (self-prepared, powder from Americanbio, catalog number: CU01100-00200)
IPTG, 500 mM (self-prepared, powder from TEKnova, catalog number: I3305)
Phenylmethanesulfonyl fluoride (PMSF), 1 M (self-prepared, powder from MilliporeSigma, catalog number: 78830)
HisTrap HP 5 mL column (Cytiva, catalog number: 17524801)
10%–20% Tris-Glycine SDS-PAGE gel (Thermo, Invitrogen, catalog number: XP10205BOX)
Tris-glycine SDS running buffer, 10× (Thermo, Invitrogen, catalog number: LC2675)
Protein ladder (New England Biolabs, catalog number: P7717S)
Blue protein loading dye with reducing reagent (New England Biolabs, catalog number: B7703S)
DTT (MilliporeSigma, catalog number: D9779)
SimplyBlue SafeStain reagent (Thermo, Life Technologies, catalog number: LC6065)
HiTrap Q 5 mL column (Cytiva, catalog number: 17115401)
Bradford protein assay kit (Thermo, catalog number: PI23200)
Dialysis tubing, 6–8 kD (VWR, catalog number:28170-138)
LB agar plate containing 40 μg/mL kanamycin (self-prepared)
LB media (see Recipes)
Iron II solution (see Recipes)
Carbamoylphosphate solution (see Recipes)
TE buffer (see Recipes)
Cell lysis buffer (see Recipes)
HisTrap column buffer A (see Recipes)
HisTrap column buffer B (see Recipes)
HiTrap Q column buffer A (see Recipes)
HiTrap Q column buffer B (see Recipes)
Dilution buffer (see Recipes)
Dialysis buffer (see Recipes)
Ultrapure EDTA, 0.5 M, pH 8.0 (ThermoFisher, catalog number: 15575020), store at RT
Tris-HCl, 1 M, pH 8.0 (Amresco, catalog number: E298-NEB-2L), store at RT
Tris-HCl, 1 M, pH 7.5 (Amresco, catalog number: E199-NEB-2L), store at RT
NaCl (MilliporeSigma, catalog number: 203505), store at RT
Glycerol (self-prepared), store at RT
Imidazole (MilliporeSigma, catalog number: I2399), store at RT
Tryptone (MilliporeSigma, catalog number: T7293), store at RT
Yeast extract (MilliporeSigma, catalog number: Y1625), store at RT
Nuclease-free water (New England Biolabs, catalog number: B1500), store at RT
Ammonium iron (ii) sulfate hexahydrate, 500 μM (MilliporeSigma, catalog number: 203505), store at RT
For carbamoyltransferase enzyme assay:
Phage T4gt genomic DNA (self-prepared), store in TE buffer at -20 °C
Note: E. coli bacteriophage T4 is known to have cytosine glucosylation and hydroxymethylation in its genome (Flodman et al., 2020). T4gt is a mutant deficient in DNA glucosyltransferase, and thus its genomic DNA contains 5-hydroxymethylcytosine. Up to 95% of cytosines in the T4gt genomic DNA are replaced by 5-hydroxymethylcytosine. The propagation of T4gt was done using T7 express (NEB, catalog number: C2566), and the bacterial host was grown in LB medium at 37 °C. The host was infected with phage at the log phase until the culture reached lysis. The phages in supernatant were pelleted by 10% PEG8000 and centrifugated. The genomic DNA of T4gt phage was extracted using phenol/chloroform.
5hmC DNA oligo: 5’-TGTCCGATAGACT/5dhmC/TACGCA (IDT, 5’-TGTCCGATAGACT/i5HydMe-dC/TACGCA), store at -20 °C
5hmC RNA oligo: 5’-GGAGTGAGAAGATGGT/5hmC/TAGGTGTTTATTGGTGATGAA (synthesized with in vitro transcription), store at -20 °C
5-hydroxymethyl dCTP, 100 mM (Zymo Research, catalog number: D1045), store at -20 °C
5-hydroxymethyl CTP, 100 mM (Trilink Biotechnologies, catalog number: N-1087), store at -20 °C
5’-hydroxymethylcytosine Carbamoyltransferase, store at -20 °C
Note: Please see Procedure Section A for expression and purification of the enzyme.
Proteinase K (New England Biolabs, catalog number: P8107), store at -20 °C
Thermolabile Proteinase K (New England Biolabs, catalog number: P8111), store at -20 °C
NEBuffer 2.1, 10× (New England Biolabs, catalog number: B7202), store at -20 °C
Ammonium iron (ii) sulfate hexahydrate, 500 μM (MilliporeSigma, catalog number: 203505, see recipe section), store at RT
Note: Iron (ii) solution should be freshly prepared for the experiment
Carbamoylphosphate, 100 mM (MilliporeSigma, catalog number: C4135, see Recipes), store at -20 °C
Note: Carbamoylphosphate solution should be freshly prepared for the experiment.
ATP, 100 mM (New England Biolabs, catalog number: N0450), store at -20 °C
Nuclease-free water (New England Biolabs, catalog number: B1500), store at RT
1× TE buffer (self-prepared, see Recipes), store at RT
Tris-HCl, 1 M, pH 8.0 (Amresco, catalog number: E298-NEB-2L), store at RT
Tris-HCl, 1 M, pH 7.5 (Amresco, catalog number: E199-NEB-2L), store at RT
NaCl, 5 M (self-prepared), store at RT
Glycerol (self-prepared), store at RT
Ultrapure EDTA, 0.5 M, pH 8.0 (ThermoFisher, catalog number: 15575020), store at RT
Zymo oligo clean & concentrator kit (Zymo Research, catalog number: D4060)
Norgenbiotek oligo clean-up and concentrator kit (Norgenbiotek, catalog number: 34100)
Nucleoside Digestion Mix (New England Biolabs, catalog number: M0649)
Equipment
Thermal cycler (Bio-Rad, T100, catalog number: 1861096)
Fast protein liquid chromatography (FPLC) system (GE Healthcare, model: AKTA GO)
Gel electrophoresis power supply (Thermo, model: EC105)
SDS-PAGE gel electrophoresis system (Invitrogen, model: Xcell surelock mini-cell, catalog number: EI0001)
Gel photography system (AlphaImager HP)
Sonicator for E. coli cell disruption (Misonix, model: S-4000)
Bacteria incubation shaker (New Brunswick Scientific, model: Innova 44)
Agar plate incubator at 37 °C (Napco, model: 310)
UV/Visible spectrophotometer (Amersham Biosciences, model: Ultrospec 2100 pro)
Centrifuge (Beckman Coulter, model: Avanti J-E)
Table-top centrifuge (Eppendorf, model: 5415D)
Vortex shaker
Water bath or heat block
Ice bath
Pipette
Procedure
Expression and purification of His-carbamoyltransferase protein
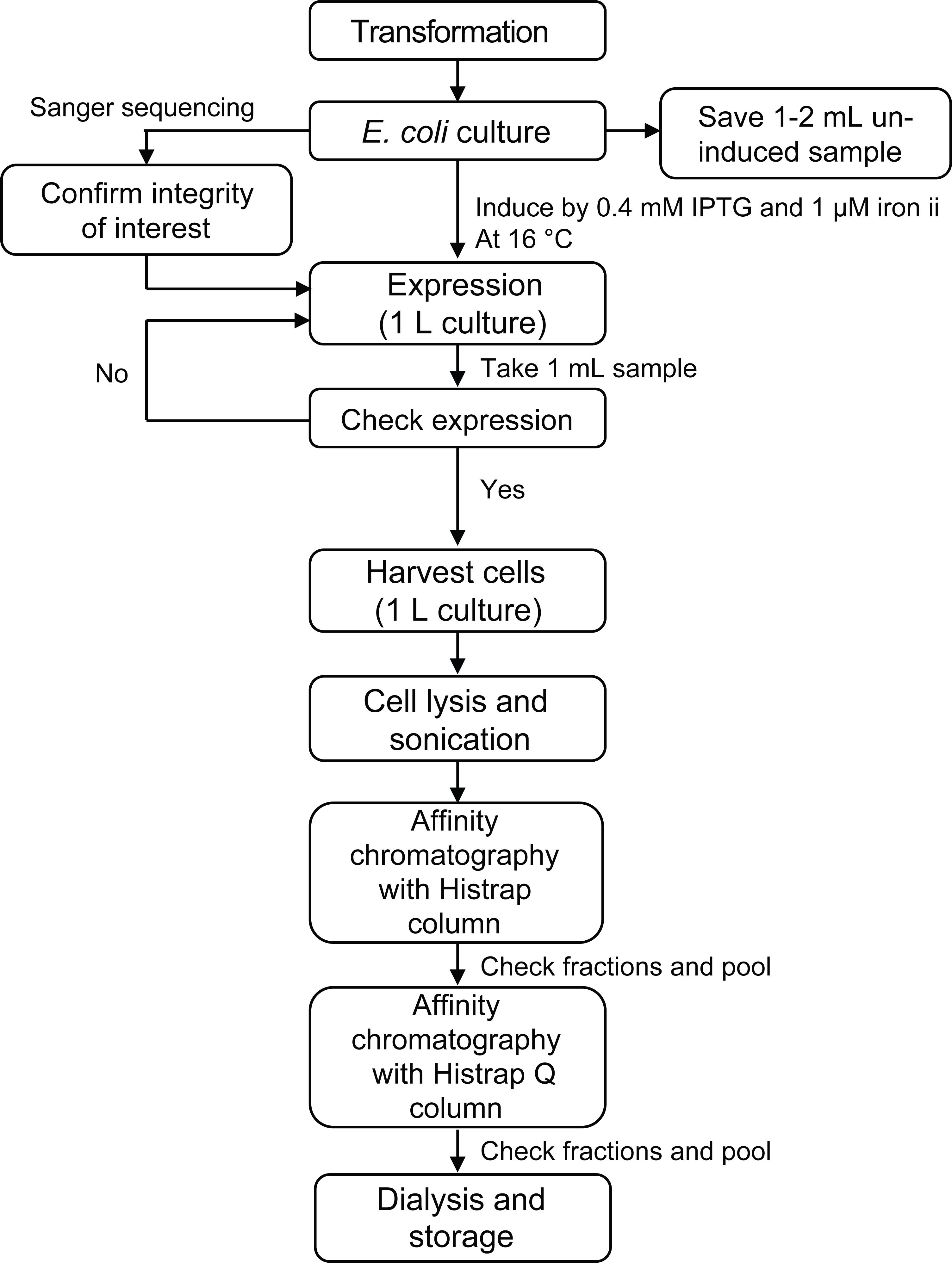
Figure 1. Major steps for expression and purification of protein.Transform 50 μL of competent DH5-alpha cells with 10–50 ng of pET28-Kan-his-CT plasmid (follow the manufacturer protocol) and spread 20–30 μL on an agar LB plate containing 40 μg/mL kanamycin. Incubate the plate overnight at 37 °C.
Inoculate 10–50 mL of LB medium containing 40 μg/mL kanamycin in a bacteria culture tube or shaking flask with a single colony from the plate. Grow overnight at 37 °C while shaking at 250 rpm.
Optional: Verify the integrity of the insert by sanger sequencing.
In the next morning, inoculate 1 L of LB medium containing 40 μg/mL kanamycin in a shaking flask with 10–25 mL overnight culture (roughly 1:40 to 1:100 ratio). Grow the cells at 37 °C while shaking at 250 rpm until OD600nm reaches 0.6. Save 1–2 mL cell culture as un-induced control. Pellet the 1–2 mL cells by centrifuging for 5 min at 10,000 × g and freeze in -20 °C for short-term storage.
Note: This step may take several hours. Check OD600nm every 1–1.5 h in the first few hours and then more frequently. Be aware that the cells grow fast in log phase and the OD600nm can increase rapidly when near 0.6.
Transfer the cells to a shaker pre-chilled to 16 °C and let it sit for 20 min to cool down the cell culture.
Note: This step is performed to guarantee that carbamoyltransferase protein expression is induced at its optimal temperature (~16 °C).
Add IPTG at 0.4 mM (stock concentration is 0.4 M, 1,000×) and 1 μM iron II (stock concentration is 500 μM, 500×) to the culture, and continue growth at 16 °C while shaking at 250 rpm for ~24 h. Carbamoyltransferase protein expression is confirmed in step A6, before the entire 1 L volume of culture is processed in step A7.
Note: Iron (ii) is added to facilitate protein folding.
Check protein expression with SDS-PAGE gel.
Harvest 1 mL culture, and pellet cells by centrifuging for 5 min at 10,000 × g.
Note: The purpose of taking 1 mL from the 1 L culture is for the confirmation of protein expression before the entire 1 L culture is harvested and processed. Note that this 1 mL culture is induced, and the 1 mL control culture taken in step A3 is un-induced.
Resuspend the pellet and un-induced cell pellets (saved in step A3) in 1 mL cell lysis buffer.
Lyse the cells in a 1.5 mL microcentrifuge tube on ice with 3–5 cycles of 20 s on/20 s off pulses using sonicator with a microtip. 10 μL is saved as crude lysate. The rest of the volume (1 mL – 10 μL = 990 μL) is used in step A6d.
Pellet the inclusion bodies by centrifuging the sonicated cell suspension at full speed for 10 min at 4°C, and transfer supernatant to a new 1.5 mL microcentrifuge tube.
Mix 10 μL crude lysate taken in step A6c, 10 μL nuclease-free water, and 10 μL blue protein loading dye (3× with reducing reagent added) to get a final volume of 30 μL.
Mix 10 μL of soluble samples (supernatant from step A6d), 10 μL of nuclease-free water, and 10 μL of blue protein loading dye (3× with reducing reagent added) to get a final volume of 30 μL.
Heat samples from steps 6Ae and 6Af at 95 °C for 5 min.
Assemble the SDS-PAGE gel and gel running apparatus, fill with 1× running buffer, load the samples and protein ladder, and run gel at 150 V for approximately 1 h.
Stain the gel with SimplyBlue staining reagent, take picture of the gel, and confirm expression of carbamoyltransferase protein in crude lysate and soluble portion.
Note: The estimated size of carbamoyltransferase enzyme is 63kDa. Presence of protein in soluble lysate should be confirmed before large-scale purification in the next step.
Harvest cells from 1 L cell culture from step A5 by centrifuging for 15 min at 5,000 × g at 4 °C.
Cell resuspension and sonication:
Directly resuspend cell pellet in 30 mL of HisTrap buffer A (or 4 mL buffer per gram of pellet).
Sonicate the cells on ice with 5 s on/5 s off pulses for 30–40 cycles. Set power to max level.
Note: The number of cycles needed for complete disruption of cells depends on the model of sonicator. It is suggested to do a pre-test to determine the best sonication condition before performing the actual experiment.
Centrifuge the sonicated cell suspension for 30 min at 23,000 × g at 4 °C.
Transfer supernatant (~30 mL) and mix with equal volume of HisTrap buffer A for loading to the AKTA purifier. Add the appropriate volume of 1M PMSF to a final concentration of 0.4 mM to prevent protein degradation.
Note: Save 0.1 mL sample as before purification control. Here, the total loading volume is about 60 mL, and the suggested loading volume range is between 50 and 100 mL.
First purification using HisTrap HP 5 mL column on AKTA.
Equilibrate the column with 10 column volume (CV) of HisTrap buffer A.
Load sample at 2 mL/min.
Wash out unbound proteins of the column using 10× CV HisTrap buffer A.
Elute protein using a linear gradient from 0%–100% HisTrap buffer B for a total of 10× CV. Set fraction volume to collect 2 mL per fraction tube.
Check purified proteins in each fraction by adding 5 μL of sample with 15 μL of nuclease-free water and 10 μL of blue protein loading dye (3×, reducing agent added) to get a final volume of 30 μL.
Heat samples at 95 °C for 5 min.
Load the samples and run SDS-PAGE gel (see Figure 2 for example of protein purification with HisTrap column).
Stain the gel with SimplyBlue staining reagent, and take picture of the gel.
Pool fractions with concentrated proteins.
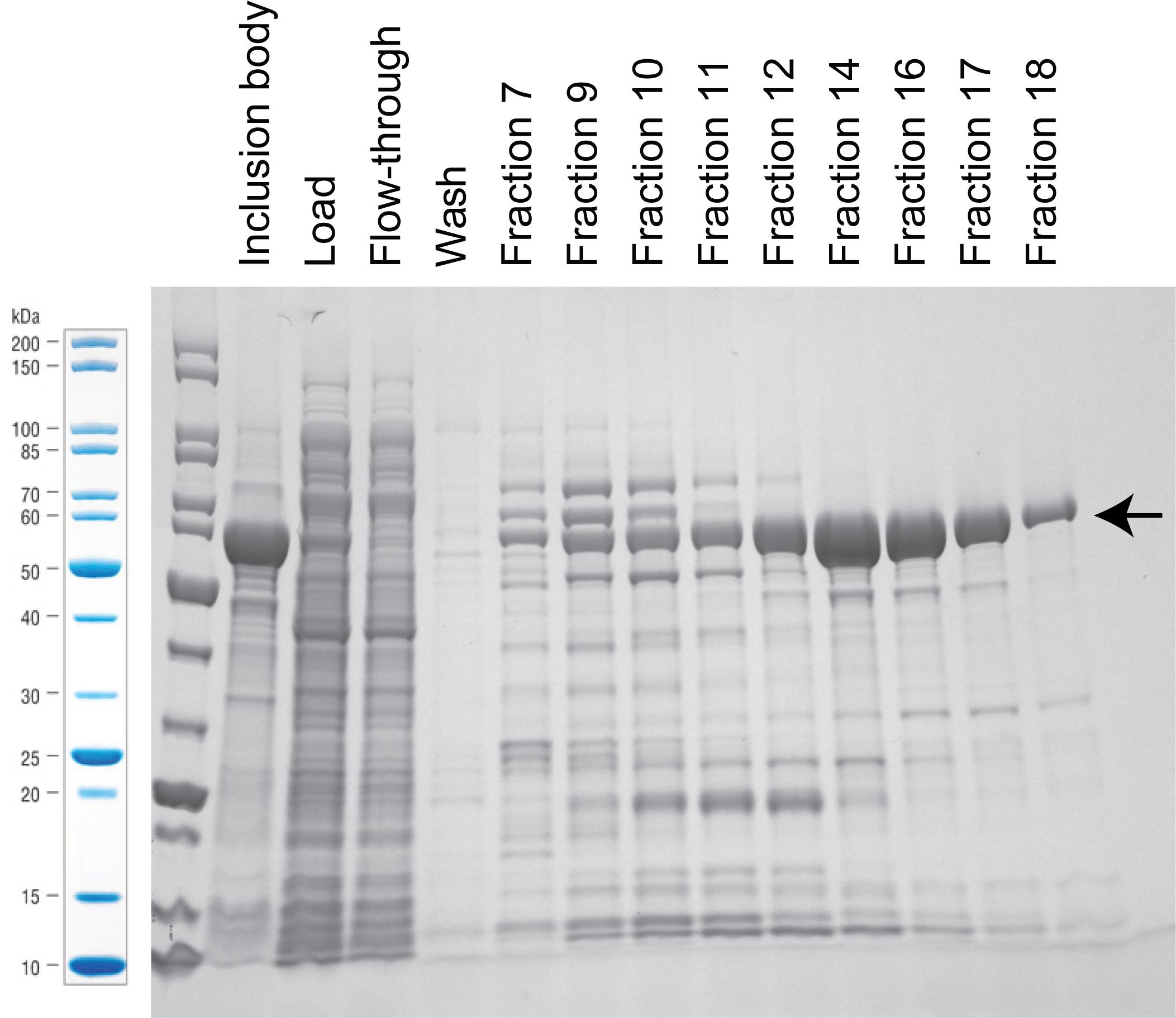
Figure 2. Stained SDS-PAGE gel with samples from the purification with HisTrap column. Arrow marks band of carbamoyltransferase expression at 63 kDa. For this particular example, we pooled fraction 12–18.Second purification using HiTrap Q column on AKTA.
Mix pooled fractions with 7× volume of dilution buffer so that the final salt concentration is approximately 50 mM.
Equilibrate the column with 10 column volume (CV) of Q buffer A.
Load sample at 3 mL/min.
Wash out unbound proteins of the column using 10× CV Q buffer A.
Elute protein using a linear gradient from 0%–100% Q buffer B for a total of 10× CV. Set fraction volume to collect 2 mL per fraction tube.
Check purified proteins in each fraction by adding 5 μL of sample with 15 μL of nuclease-free water and 10 μL of blue protein loading dye (3×, reducing agent added) to get final volume of 30 μL.
Heat samples at 95 °C for 5 min.
Load the samples and run SDS-PAGE gel (see Figure 3 for example of protein purification with HiTrap Q column).
Stain the gel with SimplyBlue staining reagent (following manufacturer instructions), and take picture of the gel.
Pool fractions with concentrated proteins.
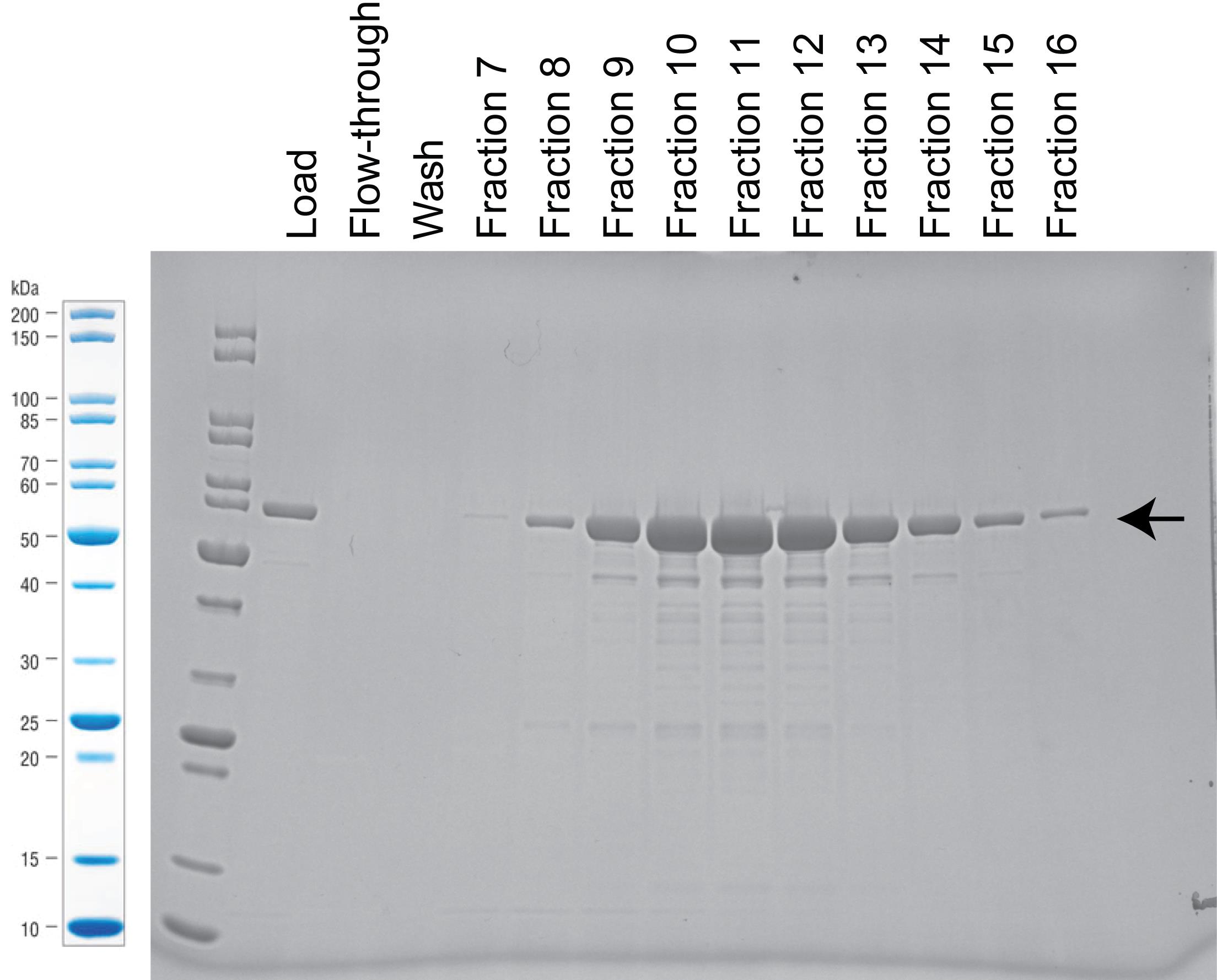
Figure 3. Stained SDS-PAGE gel with samples from the purification with Hitrap Q column. Arrow marks band of carbamoyltransferase expression at 63 kDa. For this particular example, we pooled fraction 9–13.
Transfer purified protein to a dialysis tubing bag (MWCO: 6–8 kD). Dialyze overnight against dialysis buffer at 4 °C.
Carefully transfer samples from dialysis tubing bag and determine concentration of purified carbamoyltransferase using the Bradford protein assay kit (following manufacturer instructions).
Aliquot purified carbamoyltransferase into 50 μL per microcentrifuge tube and store at -20 °C.
dsDNA enzymatic assay
DNA denaturation: 5-hydroxymethylcytosine carbamoyltransferase has a preference for ssDNA; therefore, it is critical to denature dsDNA to ssDNA for optimal conversion.
Prepare 1–2 μg genomic DNA in 5 μL of nuclease-free water or TE buffer in PCR tube
Note: The reaction with T4gt genomic DNA (containing 90–90% 5hmC) is shown as example.
Heat to denature dsDNA in a thermal cycler set at 95 °C for 10 min.
Immediately transfer the tube into an ice bath to avoid re-annealing.
Critical: DNA denaturation is a reversible process. When the temperature is cooled down slowly, denatured ssDNA will anneal again. Thus, it is critical to transfer the heated sample and immediately embed the tube completely in the ice bath.
Add reaction mix (see Table 1) to denatured DNA sample, keep the tube on ice, and mix thoroughly by pipetting up and down for 10 times.
Table 1. Reaction mix for dsDNA modification
Reagent Stock Conc. Final Conc. 50 μL Rxn. 5hmC denatured DNA - 1–2 μg 5 μL NEBuffer 2.1 10× 1× 5 μL Iron II solution 500 μM 10 μM 1 μL Carbamoylphosphate2 100 mM 10 mM 5 μL ATP 100 mM 5 mM 2.5 μL Enzyme - 1–7 μM See footnote1 Nuclease-free water - - Bring up to 50 μL Total - - 50 μL 1 The volume of enzyme added to the reaction should not exceed 10% of the total volume.
2 Carbamoylphosphate has a short half-life at room temperature. Therefore, it is recommended to always keep the solution on ice and to add the enzyme right before the incubation step.
Incubate the reaction in a thermal cycler set to 30 °C for 3 h.
Inactivate the reaction by adding 2 μL of Proteinase K, mix thoroughly, and continue incubating at 37 °C for 30 min.
Follow the Zymo oligo clean & concentrator kit manual to purify DNA product, resuspend, and recover DNA in 20 μL TE buffer or Elution buffer provided in the kit.
After purification, the conversion rate can be measured by quantitative liquid chromatography (see Data analysis).
Use the purified reactions for downstream analysis, or otherwise store at -20 °C.
ssDNA oligo and RNA oligo enzymatic assay
Prepare 5hmC ssDNA or RNA oligo: no denaturation is needed for single-stranded oligos; just prepare 1 μg of oligos in suitable volume between 1–5 μL with nuclease-free water or TE buffer.
Note: Examples of ssDNA or RNA oligo sequences are provided in the Materials and Methods section.
Add reaction mix (see Table 2) to oligo sample, keep the tube on ice, and mix thoroughly by pipetting up and down 10 times.
Table 2. Reaction mix for ssDNA or RNA modification
Reagent Stock Conc. Final Conc. 50 μL Rxn. ssDNA/RNA oligo - 1 μg 1–5 μL NEBuffer 2.1 10× 1× 5 μL Iron II solution 500 μM 10 μM 1 μL Carbamoylphosphate2 100 mM 10 mM 5 μL ATP 100 mM 5 mM 2.5 μL Enzyme - 1–7 μM See footnote1 Nuclease-free water - - Bring up to 50 μL Total - - 50 μL 1 The volume of enzyme added to the reaction should not exceed 10% of the total reaction volume.
2 Carbamoylphosphate has a short half-life at room temperature. Therefore, it is recommended to always keep the solution on ice and to add the enzyme right before the incubation step.
Incubate the reaction in a thermal cycler set to 30 °C for 3 h.
Inactivate the reaction by adding 2 μL of Proteinase K, mix thoroughly, and continue incubating at 37 °C for 30 min.
For purification of short oligos ≤21 nt, we generally see better recovery with Norgenbiotek Oligo clean-up and concentrator kit. Otherwise, follow the Zymo oligo clean & concentrator kit manual. Resuspend and recover converted DNA/RNA oligo in 20 μL of TE buffer or Elution buffer provided in the kit.
After purification, the conversion rate can be measured by quantitative liquid chromatography (see Data analysis).
Use the purified reactions for downstream analysis, or otherwise store at -20 °C.
Single nucleotide/deoxynucleotide enzymatic assay
Prepare 1:10 dilution of dhmCTP or hmCTP (dilute to 10 mM) with nuclease-free water.
Add reaction mix (see Table 3) to sample, keep the tube on ice, and mix thoroughly by pipetting up and down 10 times.
Table 3. Reaction mix for dhmCTP or hmCTP modification
Reagent Stock Conc. Final Conc. 50 μL Rxn. dhmCTP or hmCTP 10 mM 0.2 mM 1 μL NEBuffer 2.1 10× 1× 5 μL Iron II solution 500 μM 10 μM 1 μL Carbamoylphosphate2 100 mM 10 mM 5 μL ATP 100 mM 5 mM 2.5 μL Enzyme - 1–7 μM See footnote1 Nuclease-free water - - Bring up to 50 μL Total - - 50 μL 1 The volume of enzyme added to the reaction should not exceed 10% of total reaction volume.
2 Carbamoylphosphate has a short half-life at room temperature. Therefore, it is recommended to always keep the solution on ice and to add the enzyme right before the incubation step.
Incubate the reaction in a thermal cycler set to 30 °C for 3 h.
Inactivate the reaction by adding 2 μL of Proteinase K, mix thoroughly, and continue incubating at 37 °C for 30 min. Because no purification will be performed to clean up Proteinase K, if it remains a concern for downstream analysis, use thermolabile proteinase K instead and perform additional heat inactivation at 55 °C for 10 min.
After the reaction, the conversion rate can be measured by quantitative liquid chromatography (see Data analysis).
Use the reactions for downstream analysis, or otherwise store at -20 °C.
The optimal concentration of enzyme for the assay depends on the active units of purified enzyme, and thus can vary from batch to batch. We recommend conducting enzyme serial dilutions to first determine the optimal molarity concentration for the reaction. For calculation of enzyme molarity, use the formula: Molarity = Moles of enzyme/volume of reaction = Mass/(Molecular weight * volume of reaction). The molecular weight of carbamoyltransferase is 63,000 g/mol.
For dsDNA enzymatic assay, we normally see up to 70% conversion using the T4gt genomic DNA; the conversion of 5dhmC to 5dcmC on ssDNA (sequence provided in Materials and Reagents) can reach nearly 100% (example data shown in Data analysis; we suggest using purification batch with ssDNA conversion rate no less than 95%); the conversion for single nucleotide/deoxynucleotide is expected to be between 60%–90%.
LB media
Weigh 10 g of tryptone, 5 g of yeast extract, and 10 g of sodium chloride.
Dissolve in 1 L H2O.
Sterilize by autoclaving.
Iron II solution (prepare fresh, this recipe makes 50 mL)
Weigh 10 mg of ammonium iron (ii) sulfate hexahydrate powder.
Dissolve in 5 mL of nuclease-free water to get 5 mM.
Dilute 1:10 with nuclease-free water to get stock concentration at 500 μM.
Carbamoylphosphate solution (prepare fresh)
Weigh 18 mg of carbamoyl phosphate powder.
Dissolve in 1 mL of nuclease-free water to get stock concentration at 100 mM.
TE buffer
Reagent Final concentration Amount EDTA (0.5 M, pH 8.0) 1 mM 0.2 mL Tris-HCl (1 M, pH 8.0) 10 mM 1 mL Nuclease-free water n/a 98.8 mL Total n/a 100 mL Cell lysis buffer
Reagent Final concentration Amount NaCl (5 M) 250 mM 5 mL Tris-HCl (1 M, pH 7.5) 20 mM 2 mL Nuclease-free water n/a 93 mL Total n/a 100 mL HisTrap Buffer A
Reagent Final concentration Amount NaCl (5 M) 250 mM 25 mL Tris-HCl (1 M, pH 7.5) 20 mM 10 mL Imidazole (2.5 M, pH 8.0) 20 mM 4 mL Nuclease-free water n/a 461 mL Total n/a 500 mL HisTrap Buffer B
Reagent Final concentration Amount NaCl (5 M) 500 mM 30 mL Tris-HCl (1 M, pH 7.5) 20 mM 6 mL Imidazole (2.5 M, pH 8.0) 750 mM 90 mL Nuclease-free water n/a 174 mL Total n/a 300 mL Dilution buffer
Reagent Final concentration Amount Tris-HCl (1 M, pH 7.5) 20 mM 6 mL Glycerol 5% 15 mL Nuclease-free water n/a 279 mL Total n/a 300 mL HiTrap Q Buffer A
Reagent Final concentration Amount NaCl (5 M) 50 mM 5 mL Tris-HCl (1 M, pH 7.5) 20 mM 10 mL Glycerol 5% 25 mL Nuclease-free water n/a 460 mL Total n/a 500 mL HiTrap Q Buffer B
Reagent Final concentration Amount NaCl (5 M) 1.5 M 90 mL Tris-HCl (1 M, pH 7.5) 20 mM 6 mL Glycerol 5% 15 mL Nuclease-free water n/a 189 mL Total n/a 300 mL Dialysis buffer (same as protein storage buffer)
Reagent Final concentration Amount Glycerol 50% 500 mL Tris-HCl (1 M, pH 7.5) 20 mM 20 mL NaCl (5 M) 100 mM 20 mL DTT (1 M) 1 mM 1 mL Nuclease-free water n/a 459 mL Total n/a 1,000 mL - Ferrer, M., Martinez-Abarca, F. and Golyshin, P. N. (2005). Mining genomes and 'metagenomes' for novel catalysts. Curr Opin Biotechnol 16, 588-593.
- Flodman, K., Correa Jr, I. R., Dai, N., Weigele, P. and Xu, S. Y. (2020). In vitro Type II restriction of bacteriophage DNA with modified pyrimidines. Front Microbiol 11: 604618.
- Lehman, I. R. and Pratt, E. A. (1960). On the structure of the glucosylated hydroxymethylcytosine nucleotides of coliphages T2, T4, and T6. J Biol Chem 235: 3254-3259.
- Yang, W., Lin, Y. C., Johnson, W., Dai, N., Vaisvila, R., Weigele, P., Lee, Y. J., Correa Jr, I. R., Schildkraut, I. and Ettwiller, L. (2021). A genome-phenome association study in native microbiomes identifies a mechanism for cytosine modification in DNA and RNA. Elife 10: 70021.
Data analysis
Quantification of the enzymatic reaction is done by measuring the conversion rate of 5dhmC/5hmC to 5dcmC/5cmC using liquid chromatography. To prepare nucleosides for quantitative analysis, we use Nucleoside Digestion Mix (New England Biolabs, catalog number: M0649) and follow the manual for incubation. No additional purification is required for LC-MS analysis. LC-MS analysis was performed on an Agilent 1200 Series LC/MS system equipped with a G1315D diode array detector and a 6120 Single Quadrupole Mass Detector in both positive (+ESI) and negative (-ESI) electrospray ionization modes. LC was performed on a Waters Atlantis T3 column (4.6 × 150 mm, 3 μm) with a gradient mobile phase consisting of aqueous ammonium acetate (10 mM, pH 4.5) and methanol. The identity of each peak was confirmed by MS. The relative abundance of each nucleoside was determined by the integration of each peak at 260 nm or its respective UV absorption maxima. Since the extinction coefficient constant for 5dcmC or 5cmC was not available, the relative quantification was performed by measuring the decrease of 5dhmC or 5hmC. The total abundance of 5dhmC/5hmC and converted 5dcmC/5cmC was normalized to a no enzyme treatment control.
Figure 4 shows an example of nucleoside peaks with a reaction performed on 5dhmC ssDNA (marked in red, sequence provided in Materials and Methods). A no enzyme reaction control is also included (marked in blue). Note the complete disappearance of 5dhmC in the red sample, which suggested almost complete conversion of 5dhmC to 5dcmC. Three replicate reactions were performed for statistical analysis.
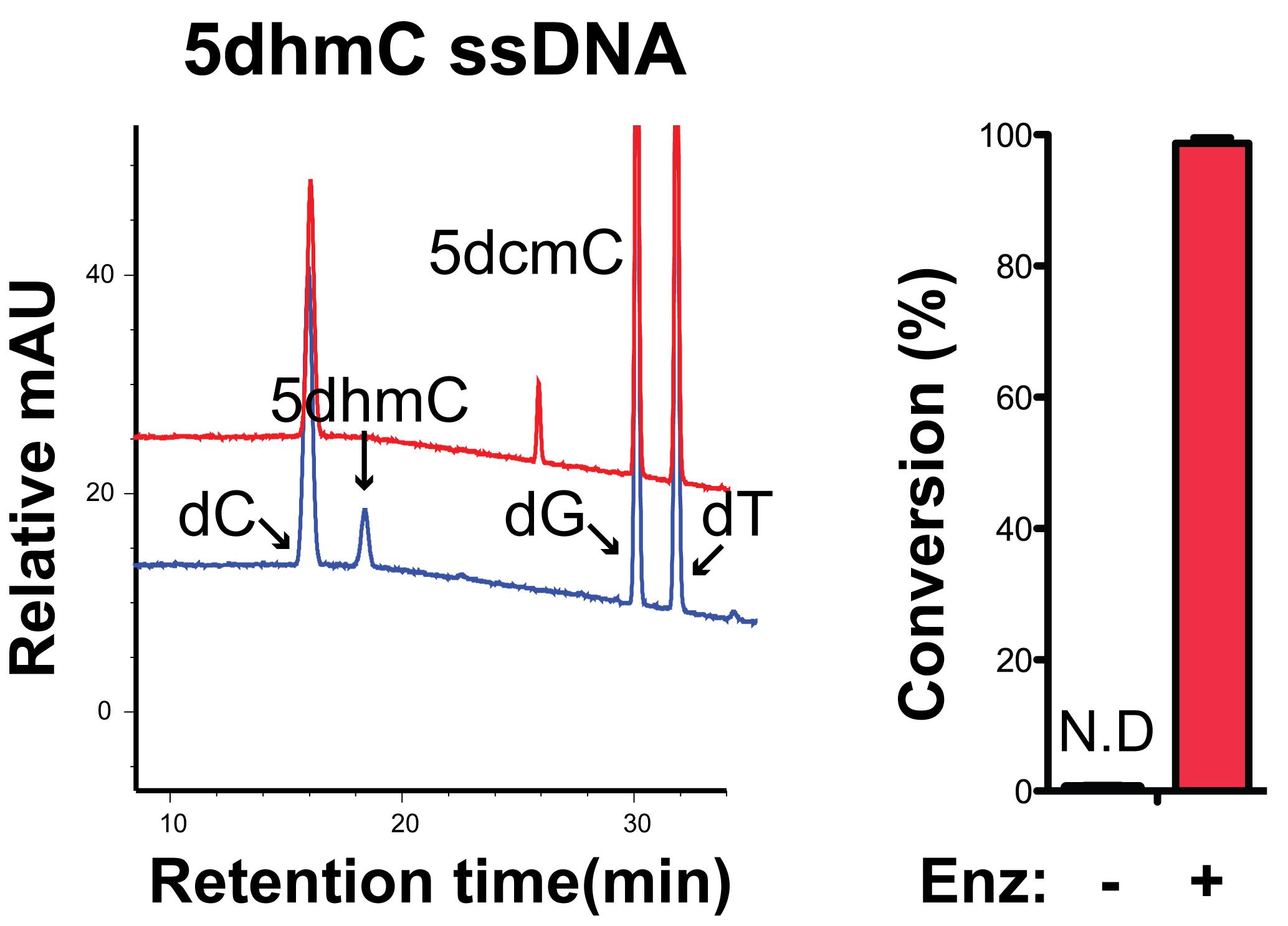
Figure 4. Quantification of carbamoyltransferase conversion rate in ssDNA oligo containing an internal 5dhmC.
Notes
Recipes
Acknowledgments
This work is funded by New England Biolabs Inc.
Competing interests
All authors are employees of New England Biolabs Inc., a manufacturer and vendor of molecular biology reagents, including several enzymes and buffers used in this work.
References
Article Information
Copyright
Yang et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Yang, W., Dai, N., Lin, Y. C., Johnson, W., Vaisvila, R., Weigele, P., Lee, Y. J., Schildkraut, I., Corrêa Jr, I. R. and Ettwiller, L. M. (2022). Carbamoyltransferase Enzyme Assay: In vitro Modification of 5-hydroxymethylcytosine (5hmC) to 5-carbamoyloxymethylcytosine (5cmC). Bio-protocol 12(17): e4496. DOI: 10.21769/BioProtoc.4496.
- Yang, W., Lin, Y. C., Johnson, W., Dai, N., Vaisvila, R., Weigele, P., Lee, Y. J., Correa Jr, I. R., Schildkraut, I. and Ettwiller, L. (2021). A genome-phenome association study in native microbiomes identifies a mechanism for cytosine modification in DNA and RNA. Elife 10: 70021.
Category
Molecular Biology > DNA > DNA modification
Biochemistry > Protein > Expression
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









