- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In-Cell Western Protocol for Semi-High-Throughput Screening of Single Clones
(*contributed equally to this work) Published: Vol 12, Iss 16, Aug 20, 2022 DOI: 10.21769/BioProtoc.4489 Views: 3474
Reviewed by: Chiara AmbrogioJohn W PetersonAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
DNA in situ Hybridizations for VEGFA Gene Locus (6p12) in Human Tumor Tissue
Mariacarla Andreozzi [...] Luigi Terracciano
Aug 5, 2015 9957 Views

Enhanced-ice-COLD-PCR for the Sensitive Detection of Rare DNA Methylation Patterns in Liquid Biopsies
Florence Mauger and Jörg Tost
Dec 5, 2019 6403 Views

Single-cell qPCR Assay with Massively Parallel Microfluidic System
Marta Prieto-Vila [...] Yusuke Yamamoto
Mar 20, 2020 5758 Views
Abstract
The in-cell western (ICW) is an immunocytochemical technique that has been used to screen for effects of siRNAs, drugs, and small molecule inhibitors. The reduced time and number of cells required to follow this protocol illustrates its semi-high-throughput nature. Performing a successful ICW protocol requires fixing and permeabilizing adherent cells directly in the plate that specifically exposes the epitope of interest. After blocking of non-specific proteins, the cells are incubated overnight with a primary antibody of interest, which is detected via a host-specific near-infrared fluorescently labeled LI-COR secondary antibody. In the final step, the plate is scanned using an Odyssey LI-COR Imaging System or similar, and each of the wells is quantified. For the first time, this technique has been demonstrated to be reproducibly utilized for semi-high-throughput selection of knockout or overexpression clones.
Graphical abstract:

Background
Knockout or overexpression screens are efficient methods for identifying the involvement of novel genes that contribute to phenotypes such as drug resistance. Perturbation of gene function is enabled through either loss-of-function studies, using biological tools such as the CRISPR-Cas9 system (Clustered Regularly Interspaced Short Palindromic Repeats) (Humphrey and Kasinski, 2015; Szlachta et al., 2018; Li and Kasinski, 2020), or by gain-of-function studies through overexpression of human ORFs (Open Reading Frame) (Arnoldo et al., 2014). Several genes can potentially be identified by such screening methodologies; however, validation is a key step. To validate and dissect the cellular function of the gene(s) of interest, gene modulation has become a prevalent technique in the field. To this end, the candidate gene is either individually knocked out or overexpressed, single clones are isolated, and finally the phenotype observed via the screen is reevaluated. Nevertheless, to confirm that the gene of interest is accurately knocked out or overexpressed in single clones, protein quantification is a usual process.
Differential protein expression of individual clones is conventionally determined by western blot. This and other techniques such as immunofluorescence and immunohistochemistry are indispensable methods for protein analysis; however, these protocols require increased amounts of resources (antibodies, reagents) and often need one or more days to complete. Therefore, a semi-high-throughput screen that allows for rapid identification of differential protein expression post-clonal selection that reduces expenses, labor, and time should be considered. The in-cell western (ICW) is a powerful, simple, and reproducible technique that is underutilized in the field. It is a cost-effective method to quantify intracellular signaling in intact cells. The ICW protocol involves fixation and immunostaining of cells and combines the specificity of a western blot with the reproducibility and throughput of an enzyme-linked immunosorbent assay (ELISA) (Boveia and Schutz-Geschwender, 2015). From previous reports, semi-high-throughput cell-based applications of ICW include: 1) identification of efficient siRNAs from libraries and 2) identification of small molecule inhibitors targeting a particular signaling pathway (Hoffman et al., 2010). Additionally, ICW has been successfully utilized for screening genotoxic drugs by quantifying the expression of γH2AX, a well-known DNA damage and repair marker (Khoury et al., 2013). Another example of the versatility of ICW for throughput experiments is during the screening of chemical libraries for compounds that modulate the intensity and duration of growth factor-induced MAPK activity, an important regulator in cancer progression (Schnaiter et al., 2014).
Here, we describe a semi-high-throughput screening mechanism using the ICW protocol for validation of single knockout or overexpression clones for a protein of interest, initially identified using the CRISPR-Cas9 screening system (Pal et al., 2020, 2022). ICW has proved to be an efficient technique for clonal selection of cells because it allows rapid analysis of numerous samples, conserving the accuracy of the quantifiable output. To the best of our knowledge, the ICW protocol described below is the first reported use of ICW for the selection of multiple single clones simultaneously.
The pros and cons of using ICW versus western blot for efficient clonal selection of cells are enlisted below (Table 1).
Table 1. Pros and cons of using in-cell western over using the more conventional technique of western blot for high-throughput selection of single clones.
| Pros | Cons |
|
|
The protocol described here is validated for one protein; therefore, optimization of various parameters may be necessary to achieve study-specific goals, described in Table 2.
Table 2. Parameters at specific steps of ICW that can be optimized to achieve high-throughput selection of single clones for study-specific goals.
| Steps in protocol | Parameters to be optimized |
| Antibody Optimization |
|
| Cells |
|
| Choice of plate |
|
| Fixation |
|
| Permeabilization |
|
Materials and Reagents
96-well clear flat-bottom polystyrene tissue-culture plates (Corning, catalog number: 3596)
15 mL Falcon tubes (Corning, Falcon®, catalog number: 352097)
Reagent reservoir nonsterile (VWR, catalog number: 89094-684)
100% methanol (Thermo Fisher, catalog number: A412-20), storage: room temperature
Distilled water, storage: room temperature
NaCl (Sigma Aldrich, catalog number: S3014-1KG), storage: room temperature
KCl (Sigma Aldrich, catalog number: P9333-1KG), storage: room temperature
KH2PO4 (Sigma Aldrich, catalog number: P9791-500G), storage: room temperature
Na2HPO4·2H2O (Sigma Aldrich, catalog number: 71643-1KG), storage: room temperature
Triton X (Sigma Aldrich, catalog number: X100), storage: room temperature
Tween-20 (Sigma Aldrich, catalog number: P9416), storage: room temperature
Primary Antibodies (varies), storage: either 4 °C or -20 °C, depending on the antibody
LI-COR Secondary Antibodies (varies), storage: 4 °C
1× PBS (see Recipes)
Permeabilizing buffer (0.2% Triton X in 1× PBS) (see Recipes)
1× Phosphate Buffered Saline Tween-20 (PBST) (see Recipes)
Equipment
LI-COR Blocking Buffer (LI-COR, Odyssey, catalog number: 927-40003)
Multichannel pipettes (Mettler-Toledo International, catalog number: L12-200XLS)
FinnpipetteTM Novus multichannel pipette (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 46300800)
Software
Image Studio Lite (LI-COR Biosciences, https://www.licor.com/bio/image-studio/)
Procedure
Plate 2,000 to 10,000 cells in single wells or duplicates in a 96-well plate, in appropriate culture medium, for 24 to 48 h prior to performing ICW.
Note: Include appropriate positive control cells and blank wells containing only medium for every assay. Cell number may need to be optimized for the cell type.
Chill 100% methanol at -20 °C for 15 min.
Remove media by flicking plate, then using a multichannel pipette to remove any residual media (Video 1).
Note: It was determined that flicking the plate on a stack of paper towels (turning the plate upside down rapidly and tapping gently) to decant the media was preferred over mechanical removal. Fewer cells were disrupted. This is especially true for less adherent cells.
Video 1. Procedure for flicking plate.Add 150 µL of methanol to each of the 96 wells and incubate for 20 min at 4 °C (without shaking).
Notes:
Methanol should be added very gently down the walls of each of the wells.
Be cautious that wells do not dry out throughout the process. A manual multichannel pipette will suffice for few wells in a 96-well plate. However, an electronic multichannel pipette is advisable for a high-throughput experiment (Video 2).
Video 2. Procedure for adding methanol slowly using electronic multichannel pipette.Flick plate to remove methanol, then using the multichannel pipette remove any residual methanol.
Note: See Video 1 in step 3 for flicking procedure.
Permeabilize cells using 150 µL of permeabilizing buffer with gentle shaking at room temperature for 30 min.
Flick plate to remove permeabilizing buffer, then using the multichannel pipette, remove any residual permeabilizing buffer.
Note: See Video 1 in step 3 for flicking procedure.
Block for 1.5 h using 50 µL LI-COR blocking buffer with gentle shaking at room temperature
Note: At this point, the plate can be stored at 4 °C overnight. Plates stored for longer than 4 days need to be checked for bacterial growth.
Flick plate to remove blocking buffer, then using the multichannel pipette, remove any residual buffer.
Note: See Video 1 in step 3 for flicking procedure.
Add 50 µL of 1:50 to 1:500 primary antibody and incubate overnight at 4 °C with gentle shaking.
Note: It is advised to use antibodies that are validated for ChIP or immunofluorescence. Each antibody will need to be optimized for concentration; typically, 1:50–1:500 dilutions are used due to differences in antibody affinity.
Carefully remove antibody. This can be accomplished by either flicking the plate or using the multichannel pipette if the antibody will be saved. For the latter, care should be taken to avoid disrupting the cells on the bottom of the well with the pipette tips. To avoid disrupting the cell monolayer, angle the plate at ~45° and position the pipette tips at the interface of the wall of the well and the bottom of the well. Slowly pipette the antibody solution into the pipette.
Note: Some primary antibodies can be saved and reused if desired. However, the number of reuses has to be determined for each antibody separately.
Add 150 µL of PBST to each well using the multichannel pipette and place plate on a shaker for 5 min at room temperature.
Remove PBST by flicking plate.
Note: See Video 1 in step 3 for flicking procedure.
Repeat steps 12 and 13 for a total of five times.
Add 50 µL of 1:800 secondary antibody, cover plate with aluminum foil to protect antibody from light, and incubate 1 h at room temperature with gentle shaking.
Note: Due to the sensitivity of the fluorescent antibodies to light, be sure to keep plate covered with aluminum foil after this step.
Carefully remove antibody. See step 11.
Add 150 µL of PBST to each well using the multichannel pipette and place plate on a shaker for 5 min at room temperature.
Carefully remove the wash buffer by flicking plate.
Repeat steps 17 and 18 for a total of five times.
Perform one wash with PBS for 5 min while shaking the plate.
Flick plate to remove PBS, blot the plate, clean the bottom of the plate, and scan.
Note: At this point, since the plate has yet to be blotted for the endogenous control, it is necessary to ensure a layer of PBS is maintained in the plate to prevent it from drying out.
In order to scan the plate on the LI-COR Biosciences software, select the “plate” setting and focus at “4.0 mm” along with resolution and quality according to preference.
Note: Focus set to 4.0 mm usually works best. However, this setting may need to be adjusted based on the manufacturer of the plate used. See Figures 1 and 2 below for location of plate and software settings.

Figure 1. Plate location on LI-COR, in this case positioned in the bottom left corner.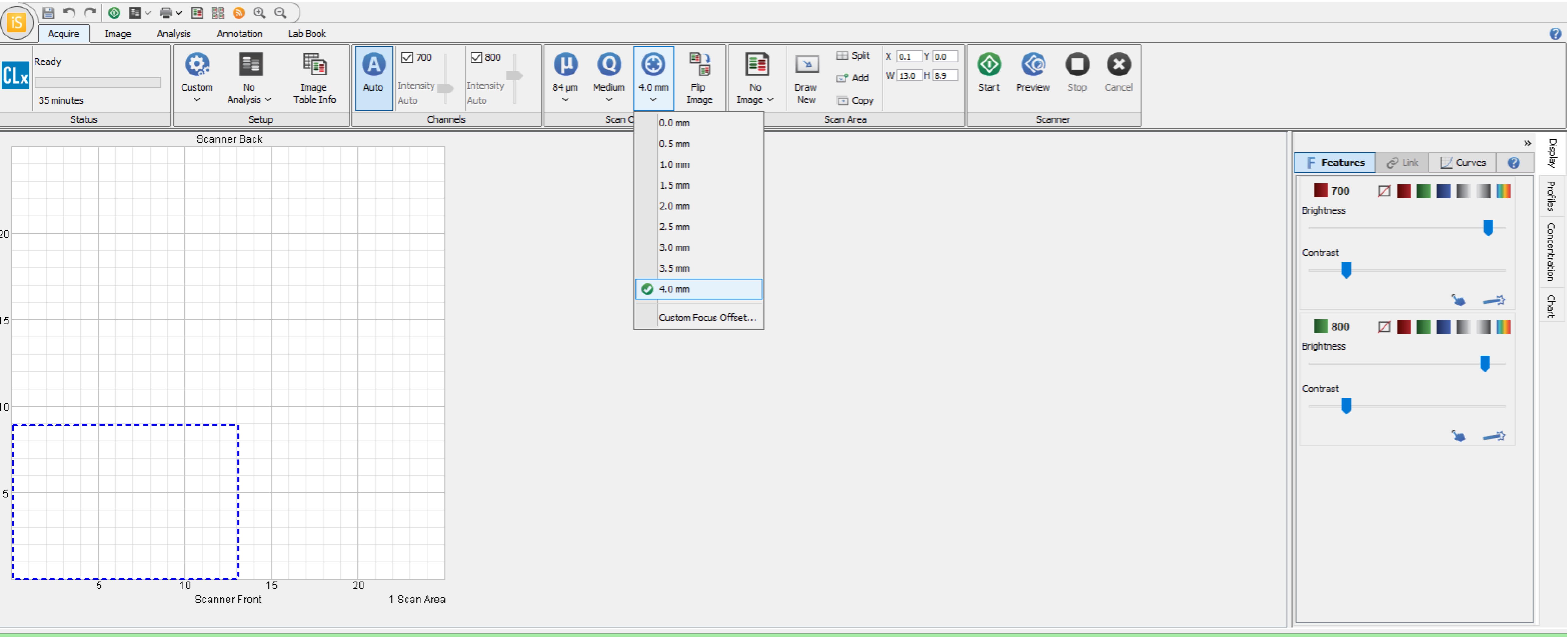
Figure 2. Plate settings on LI-COR, including selected region for scanning based on the location of the plate.Reblot the plate with the endogenous control antibody at a dilution of 1:500 to 1:1,000 either for 2.5 h at room temperature or overnight at 4 °C.
Carefully remove antibody. See step 11.
Wash each well by adding 150 µL of PBST using the multichannel pipette and incubate the plate on a shaker for 5 min at room temperature.
Repeat step 25 for a total of five times.
Add 50 µL of 1:800 secondary antibody to each well, cover the plate with aluminum foil, and incubate 1 h at room temperature with gentle shaking.
Carefully remove antibody.
Wash each well by adding 150 µL of PBST using the multichannel pipette and incubate the plate on a shaker for 5 min at room temperature.
Repeat step 29 for a total of five times.
Perform one wash with PBS for 5 min while shaking the plate.
Flick plate to remove PBS and blot the plate dry before the final scan.
Scan the plate using the suggested starting Odyssey scan parameters:
Resolution: 84 µM
Quality: Medium
Focus offset: 4.0 mm
Intensity: Adjust as necessary such that signal is evident for positive samples but not for negative controls (i.e., empty wells or cells not incubated with the primary antibody).
Note: See Figure 2 in step 22 for the setting parameters.
The specific plate format selected under the “analysis settings” tab of the software will create a grid that can be adjusted to match the wells on the scanned image for further analysis (see Figure 3 below).
Note: It is important to place the plate parallel to the scale markings on the scanner in order to align the plate template onto the scanned image.
Examples of using the ICW protocol to identify clones knocked out for a particular gene are shown in Figures 4–6. In this case, Cas9 targeting KMT5C was transfected into cells, and individual clones were isolated and propagated. Using the ICW protocol, expanded clones were then evaluated for the downstream histone modification generated of KMT5C, histone 4 lysine 20 trimethylation (H4K20me3). Clones with variability in KMT5C activity were identified using the ICW protocol (Figure 4) and were confirmed via western blotting (Figure 5). In a similar way, clones overexpressing a doxycycline-inducible KMT5C were screened using the ICW protocol (Figure 6).
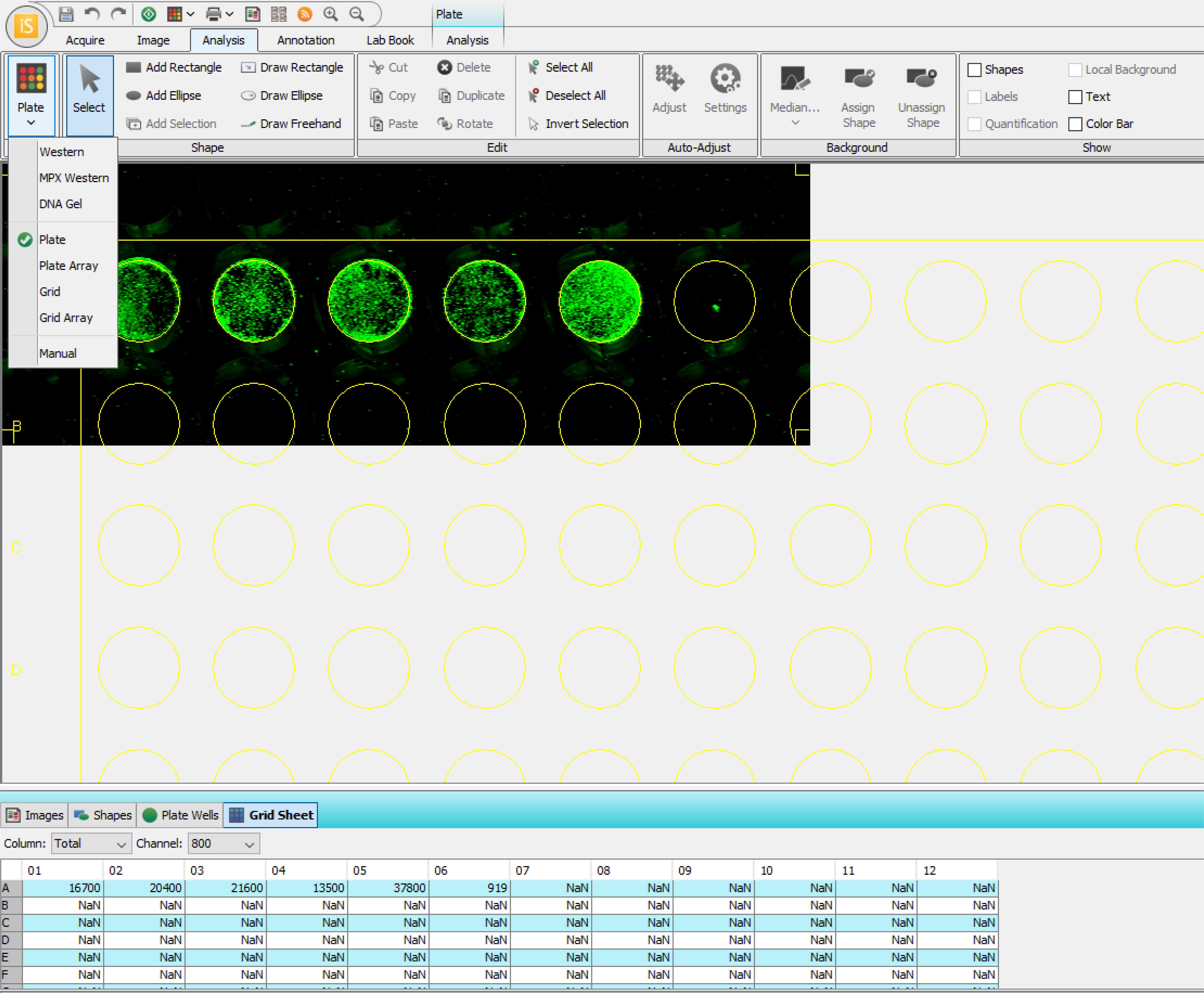
Figure 3. Image depicting the analysis setting options on the LI-COR software, including quantification of signal intensity in each identified well in the table at the bottom.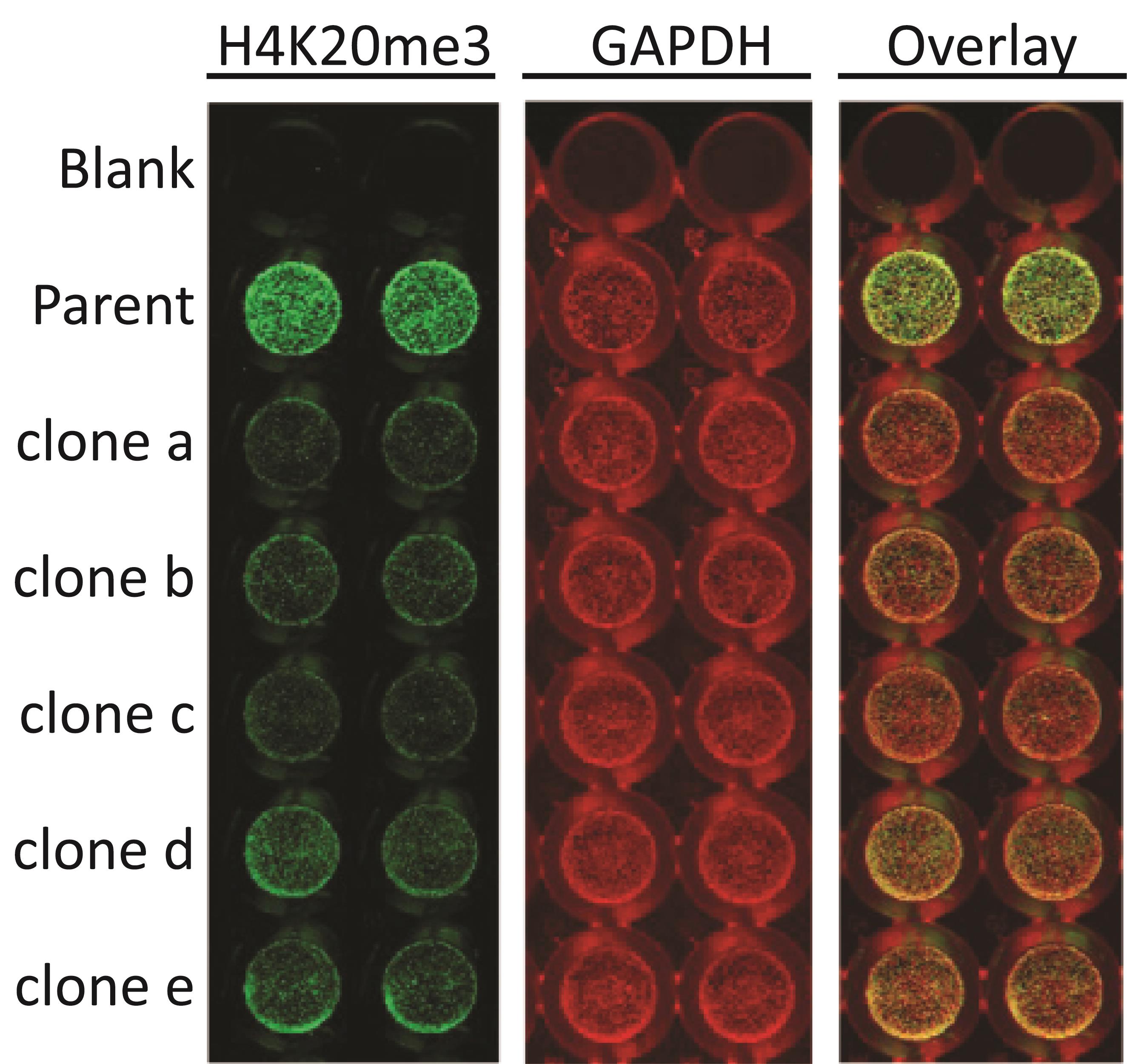
Figure 4. Selection of single clones knocked out for KMT5C by quantifying a downstream effector, H4K20me3 mark via ICW. Parent cell or KO clones (clones a-e) were plated in duplicates in a 96-well plate (10,000 cells/well). 48 h post-plating, cells were blocked using LI-COR blocking buffer, permeabilized using permeabilizing buffer, incubated with 1:400 H4K20me3 (Active Motif) primary antibody overnight on a shaker at 4 °C, and detected using anti-mouse LI-COR secondary antibodies. Then, the plate was scanned and re-blotted overnight on a shaker at 4 °C using a 1:500 concentration of GAPDH (Cell Signaling) primary antibody and detected using anti-rabbit LI-COR secondary antibodies.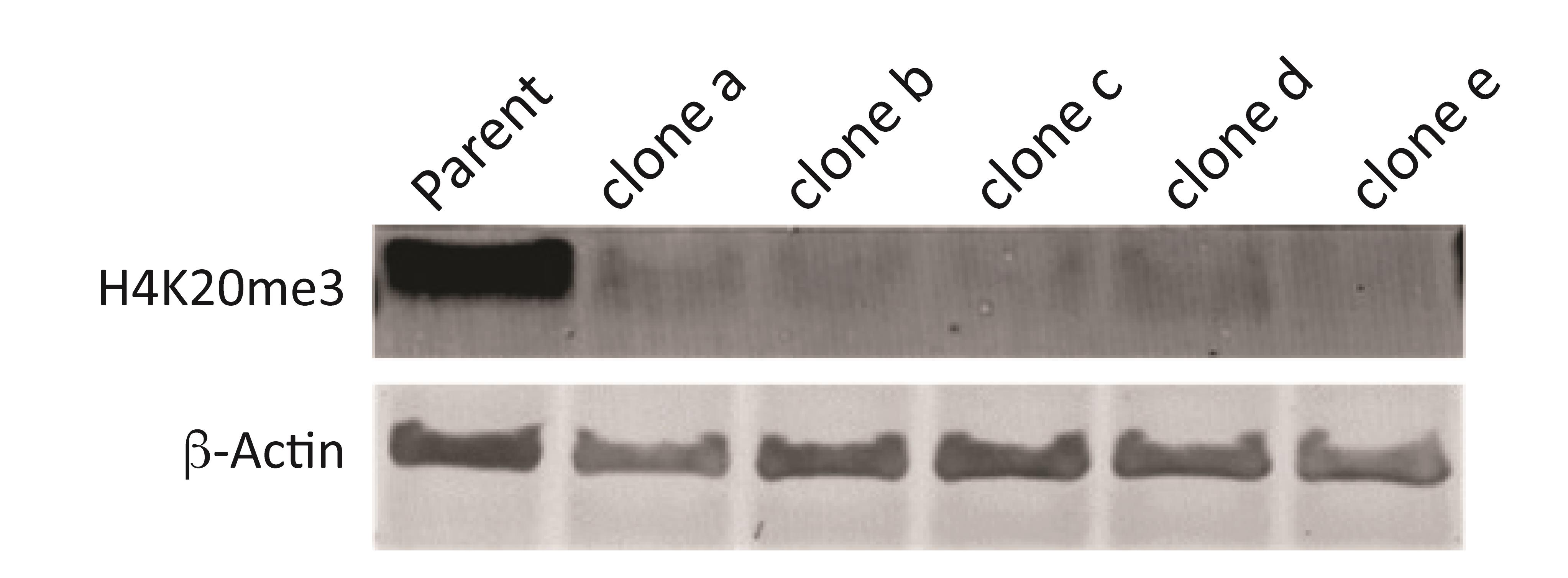
Figure 5. Clones identified through ICW, validated via western blot. Parent cells or KO clones (Clones a-e) were plated in a 6-well plate at 4 × 105 cells/well. 48 h post-plating, lysates were isolated, quantified, and separated using polyacrylamide gel electrophoresis. Post-transfer onto a PVDF membrane, the membrane was blocked using LI-COR blocking buffer and incubated overnight in 1:500 H4K20me3 antibody or in 1:10,000 β-ACTIN (Cell Signaling) primary antibody overnight on a shaker at 4 °C, and detected using anti-mouse or anti-rabbit LI-COR secondary antibodies, respectively.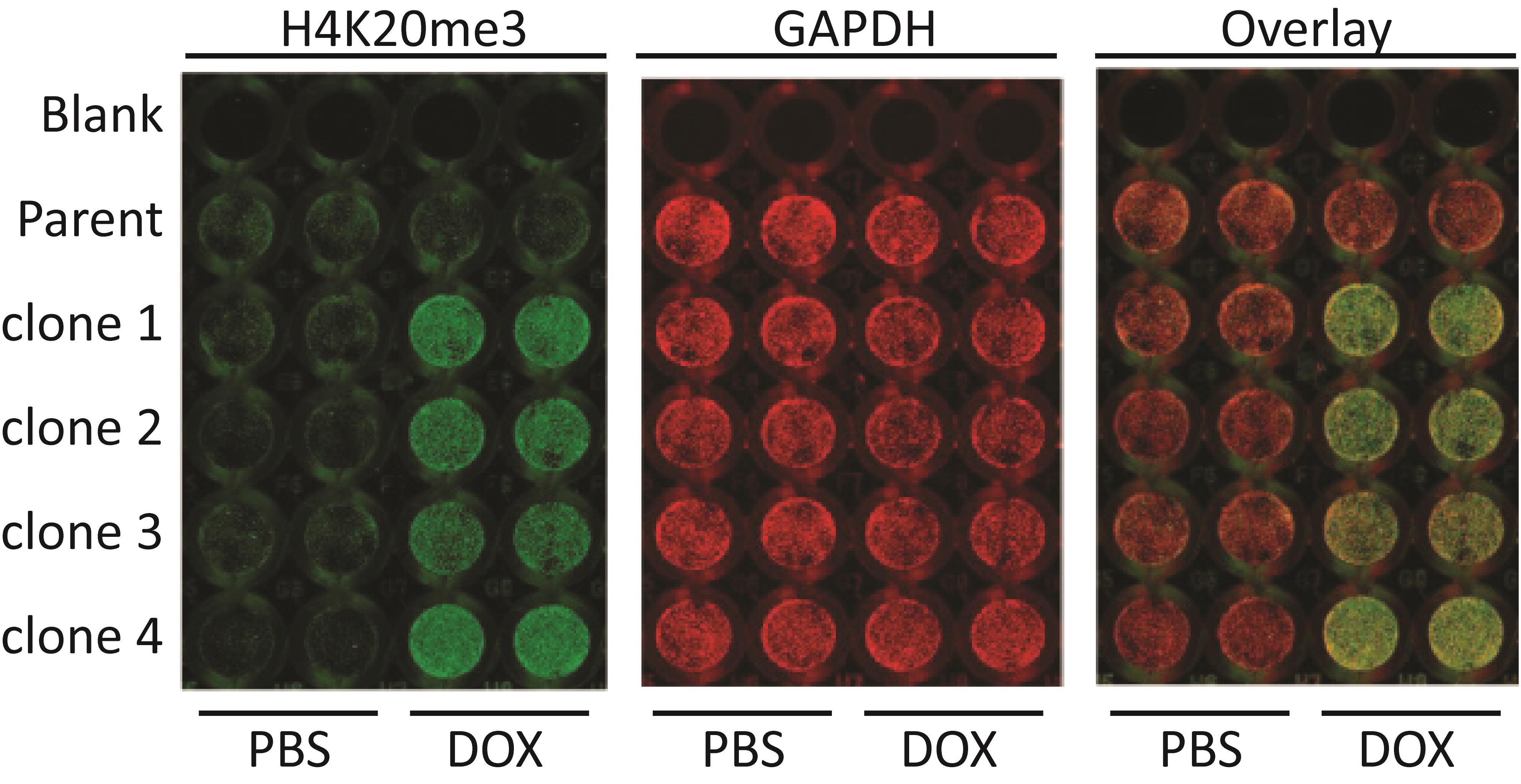
Figure 6. Single clones selected post-doxycycline mediated induction of KMT5C via H4K20me3 quantification by ICW. Either parental cells or doxycycline inducible KMT5C-single clones (clones 1–4) were plated at 10,000 cells/well in replicates of four in a 96-well plate. Doxycycline (2 µg/mL) or equivalent volume of PBS was added to two wells for each cell line at the time of plating. 48 h post-treatment, cells were blocked using LI-COR buffer, permeabilized using permeabilizing buffer, incubated with 1:400 H4K20me3 (Active Motif) primary antibody overnight on a shaker at 4 °C, and detected using anti-mouse LI-COR secondary antibodies. Then, the plate was scanned and reblotted overnight on a shaker at 4 °C in 1:500 GAPDH (Cell Signaling) primary antibody and detected using anti-rabbit LI-COR secondary antibodies.
Notes
During screening of single clones, cells can be plated in single wells, without counting. However, calculating relative signal of protein of interest to that of the positive control, post-normalizing to signal of the endogenous control is necessary.
Scanning overnight dried plates (in the dark) after blotting for both the protein of interest and the endogenous control can yield more uniform and sharper signals.
We recommend validating the candidate single clones identified through ICW via other protein quantification techniques such as western blot or immunofluorescence.
Recipes
1× PBS
800 g NaCl
20 g KCl
144 g Na2HPO4·2H2O
24 g KH2PO4
8 L of distilled water
Permeabilizing buffer (0.2% Triton X in 1× PBS)
50 mL 1× PBS
100 µL Triton X
1× PBST
1 L of 1× PBS
1 mL Tween-20
Acknowledgments
We thank Chennan Li for comments regarding use of the ICW protocol. This work was supported by National Institutes of Health NCI-R01CA205420 to A.L. Kasinski. A.S. Pal was supported by a Purdue Research Foundation (PRF) Research Grant award by the Department of Biological Sciences, Purdue University, a SIRG grant administered through the Purdue Center for Cancer Research, Purdue University, a Cancer Prevention Internship Program Graduate Research Assistantship funded by Purdue University, and a Bilsland Dissertation Fellowship awarded by the Department of Biological Sciences, Purdue University. A.M. Agredo was supported by a Ross Fellowship administered through Purdue University. R. The Graphical Abstract was created using BioRender.com. Use of this protocol has been reported in an original body of work published in Cancer Research (Pal et al., 2022).
Competing interests
The authors declare no competing financial interests.
References
- Arnoldo, A., Kittanakom, S., Heisler, L. E., Mak, A. B., Shukalyuk, A. I., Torti, D., Moffat, J., Giaever, G. and Nislow, C. (2014). A genome scale overexpression screen to reveal drug activity in human cells. Genome Med 6(4): 32.
- Boveia, V. and Schutz-Geschwender, A. (2015). Quantitative Analysis of Signal Transduction with In-Cell Western Immunofluorescence Assays. Methods Mol Biol 1314: 115-130.
- Hoffman, G. R., Moerke, N. J., Hsia, M., Shamu, C. E. and Blenis, J. (2010). A high-throughput, cell-based screening method for siRNA and small molecule inhibitors of mTORC1 signaling using the In Cell Western technique. Assay Drug Dev Technol 8(2): 186-199.
- Humphrey, S. E. and Kasinski, A. L. (2015). RNA-guided CRISPR-Cas technologies for genome-scale investigation of disease processes. J Hematol Oncol 8: 31.
- Khoury, L., Zalko, D. and Audebert, M. (2013). Validation of high-throughput genotoxicity assay screening using gammaH2AX in-cell western assay on HepG2 cells. Environ Mol Mutagen 54(9): 737-746.
- Li, C. and Kasinski, A. L. (2020). In Vivo Cancer-Based Functional Genomics. Trends Cancer 6(12): 1002-1017.
- Pal, A. S., Agredo, A. M., Lanman, N. A., Clingerman, J., Gates, K. and Kasinski, A. L. (2020). Loss of SUV420H2 promotes EGFR inhibitor resistance in NSCLC through upregulation of MET via LINC01510. bioRxiv: 2020.2003.2017.995951.
- Pal, A. S., Agredo, A., Lanman, N. A., Son, J., Sohal, I. S., Bains, M., Li, C., Clingerman, J., Gates, K. and Kasinski, A. L. (2022). Loss of KMT5C Promotes EGFR Inhibitor Resistance in NSCLC via LINC01510-Mediated Upregulation of MET. Cancer Res 82(8): 1534-1547.
- Schnaiter, S., Furst, B., Neu, J., Waczek, F., Orfi, L., Keri, G., Huber, L. A. and Wunderlich, W. (2014). Screening for MAPK modulators using an in-cell western assay. Methods Mol Biol 1120: 121-129.
- Szlachta, K., Kuscu, C., Tufan, T., Adair, S. J., Shang, S., Michaels, A. D., Mullen, M. G., Fischer, N. L., Yang, J., Liu, L., et al. (2018). CRISPR knockout screening identifies combinatorial drug targets in pancreatic cancer and models cellular drug response. Nat Commun 9(1): 4275.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Pal, A. S., Agredo Montealegre, A. and Kasinski, A. L. (2022). In-Cell Western Protocol for Semi-High-Throughput Screening of Single Clones. Bio-protocol 12(16): e4489. DOI: 10.21769/BioProtoc.4489.
Category
Cancer Biology > General technique > Molecular biology technique
Cancer Biology > General technique > Genetics > Gene expression
Molecular Biology > Protein > Detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









