- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analysis of Caenorhabditis elegans Aging-related Neurodegeneration in Chemosensory Neurons
Published: Vol 12, Iss 14, Jul 20, 2022 DOI: 10.21769/BioProtoc.4473 Views: 2655
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Alginate Gel Immobilization of Caenorhabditis elegans for Optical Calcium Imaging of Neurons
Aswathy Mangalath [...] Anoopkumar Thekkuveettil
Jun 20, 2023 2264 Views
Protocol for Inoculation of PGPR Staphylococcus sciuri to Seeds and Seedlings of Rice and Tomato Plants for Increased Root and Shoot Growth
Girija Somna [...] Dinakar Challabathula
Mar 20, 2025 2177 Views

A Novel Composite Method of Post-stroke Epilepsy Induction
Yiting Guo and Raymond Tak Fai Cheung
Jul 20, 2025 1948 Views
Abstract
Aging and neuronal deterioration constitute important risk factors for the development of neuronal-related diseases, such as different dementia. The nematode Caenorhabditis elegans has emerged as a popular model system for studying neurodegeneration diseases, due to its complete neuronal connectivity map. DiI is a red fluorescent dye that can fill the worm amphid neurons and enables the visualization of their neurodegeneration over time. This protocol provides an efficient, fast, and safe method to stain worm amphid neurons to highlight the chemosensory structures of live nematodes.
Keywords: DiIBackground
Neurodegenerative diseases share a common predisposing factor, the aging of the brain. So, a greater understanding of the normal aging brain may be necessary before we can fully understand the causes of pathological aging and cognitive decline (Yankner et al., 2008).
The existence of a complete neuronal connectivity map and genetic tractability of C. elegans make this animal model useful for studying human neurological diseases. Analysis of multiple genetic databases shows that a considerable number of human genes associated with different diseases have a significant homology to C. elegans genes, and the genetic tools available for this nematode have allowed the construction of predictive models for studying the molecular mechanism of these diseases (Alexander et al., 2014; Griffin et al., 2017).
In the aging C. elegans nervous system, synapses appear physically less robust than in younger animals. The average synapse in an elderly animal is smaller in its presynaptic density and includes fewer synaptic vesicles compared to young adults; this is similar to what occurs in the human brain, where loss of synaptic integrity contributes to its deterioration and dysfunction. Away from the synapse, typical changes in aging neurons include reduced axon caliber, poor axonal transport, smaller nuclei, and reduced cytoplasmic volume. In addition, part of the loss of mobility has an origin in defects of the nervous system (Toth et al., 2012). In recent years, numerous protocols have emerged to facilitate the visualization of C. elegans neurons, taking advantage of the transparent nature of this nematode. DiI is a long-chain dialkylcarbocyanine dye that uniformly labels neurons via lateral diffusion in the plasma membrane (Godement et al., 1987; Hofmann and Bleckmann, 1999). Neurons can be labeled either retrogradely or during dissociation. Some of the labeled membrane gradually becomes internalized and retains its fluorescence, allowing identification of cells for several weeks. DiI specifically labels amphid (ADL, ASH, ASI, ASJ, ASK, and AWB), and phasmid (PHA and PHB) IL1 and IL2 neurons, as well as IL sheath and socket cells to highlight the chemosensory structures of live nematodes (Wormatlas.org [Reference 12]). These dyes do not appear to affect the survival, development, or basic physiological properties of neurons, and do not spread detectably from labeled to unlabeled neurons. It seems likely that cells become retrogradely labeled mainly by lateral diffusion of dye in the plane of the membrane (Honig and Hume, 1986). Labeling with carbocyanine dyes has already allowed several exciting advances in developmental neurobiology (Honig and Hume, 1989). The aim of this protocol is to provide a modified technique for Dil staining, to allow the study of neurodegeneration in dementia disease models of C. elegans.
Materials and Reagents
Petri dishes 60 × 15 mm 500/cs (Fisher Scientific, catalog number: FB0875713A)
Pipette tips 2–200 µL Eppendorf® epT.I.P.S. (Eppendorf, catalog number: 022492039)
Pipette tips 50–1,000 µL Eppendorf® epT.I.P.S. (Eppendorf, catalog number: 022492055)
Eppendorf® Safe-Lock 1.5 mL microcentrifuge tubes (Eppendorf, catalog number: 022363204)
Corning® 15 mL centrifuge tubes (Corning, catalog number: 430791)
Glass media bottles 200 mL (Fisher Scientific, catalog number: FB800250)
Glass slide (25 × 75 × 0.9mm) and coverslip (22 × 22mm) (Sigma-Aldrich, catalog number: CLS294875X25-72EA; C9802-1PAK)
C. elegans strains (Caenorhabditis Genetics Center: N2 Bristol)
Escherichia coli strain OP50-1 (University of Minnesota, C. elegans Genetics Center, MN)
Bacillus subtilis strain NCIB3610 (Bacillus Genetic Stock Center, catalog numbers: 3A1 and 1A96)
1,1’-dioctadecyl-3,3,3’,3’,-tetramethylindocarbocyanine perchlorate (DiI, Sigma-Aldrich, catalog number: 468495)
Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sigma-Aldrich, catalog number: M1880)
Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P5655)
Potassium phosphate dibasic (K2HPO4) (Sigma-Aldrich, catalog number: P2222)
Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653)
Sodium azide (Sigma-Aldrich, catalog number: S8032)
NaClO Hypochlorite (Sigma-Aldrich, catalog number: 13440)
Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: S8045)
Bacto peptone (BD, BactoTM, catalog number: 211677)
Agar (Sigma-Aldrich, catalog number: A1296)
Cholesterol in absolute ethanol Cholesterol (Sigma-Aldrich, catalog number: C8667)
100% ethanol (Sigma-Aldrich, catalog number: E7023)
Calcium chloride dihydrate (CaCl2·2H2O) (Sigma-Aldrich, catalog number: C3881)
Luria broth (Sigma-Aldrich, catalog number: L3522)
M9 buffer (see Recipes)
Nematode growth medium (NGM) agar (see Recipes)
Potassium phosphate buffer (see Recipes)
1 M MgSO4 (see Recipes) 5 mg/mL cholesterol (see Recipes)
1 M CaCl2 (see Recipes)
LB media (see Recipes)
Phosphate buffer (see Recipes)
1 N NaOH (see Recipes)
Bleaching solution (see Recipes)
DiI stock solution (see Recipes)
Equipment
Fluorescence microscope Zeiss (Carl Zeiss, Serial number: 3108012846)
Shaker (Axyos Technologies, catalog number: S04036.tu)
Autoclave (Tuttnauerusa, model: 6690)
Worm pick. Worm picks can either be purchased (Genesee Scientific, catalog number: 59-AWP) or made in the lab as described in Wollenberg et al. (2013)
Centrifuge (Eppendorf, model: 5430)
Tabletop centrifuge (Eppendorf, model: 5424)
Bunsen burner (Humbolt, catalog number: H-5870)
Procedure
C. elegans maintenance
Inoculate an E. coli OP50 colony into 50 mL of LB broth in a 200 mL flask, and incubate with gentle agitation at 37°C overnight (1 × 108 UFC/mL; OD: 1–1.2).
Apply approximately 200 μL of OP50 liquid culture to a 60 mm NGM plate. Incubate seeded plates at 37°C overnight before use.
Worms can be chunked (transfer agar pieces carrying worms from an old plate to a new one every 3–4 days).
Keep worms at 20°C.
C. elegans synchronization by bleaching
Allow previously synchronized worms to grow until L4 larval stage (approximately 48 h, see Figure 1), and then adult stage (~8 more hours).
Recover gravid adults in 15-mL tubes by washing plates with M9 buffer (see Recipe 1).
Pellet worms by centrifuging at 400 × g (~1,500 rpm on a standard table centrifuge) and room temperature for 2 min, and discard supernatant.
Perform 1–3 washes until the buffer appears clear of bacteria.
Add 5 mL of bleaching solution (see Recipe 8) into the tube, and shake vigorously at room temperature for up to 5 min. Bleaching longer than 5 min will kill the eggs. We followed destruction of worm bodies under the dissecting microscope, and the reaction is stopped when traces of adults are still visible, which typically takes between 3 and 5 min.
Stop reaction by adding M9 buffer until the 15-mL mark of the conical tube.
Quickly centrifuge (since treatment may still be active) at 400 × g for 1 min, and discard the supernatant.
Wash the pellet three more times, by filling the tube with M9 buffer.
Add 1 mL of M9 buffer to the pellet, or place the eggs to unseeded NGM plates, and incubate at the desired temperature with gentle agitation. Proper aeration should be provided to obtain all animals in stage L1. We check all worms are in larval stage 1 before proceeding further.
Transfer the L1 population to NGM agar plates previously seeded with the corresponding bacterial food, and incubate until they reach the young adult stage, approximately 48 h later (see Figure 1). Most of the C. elegans strains are maintained at 20°C on NGM media seeded with E. coli OP50 or B. subtilis NCIB3610.
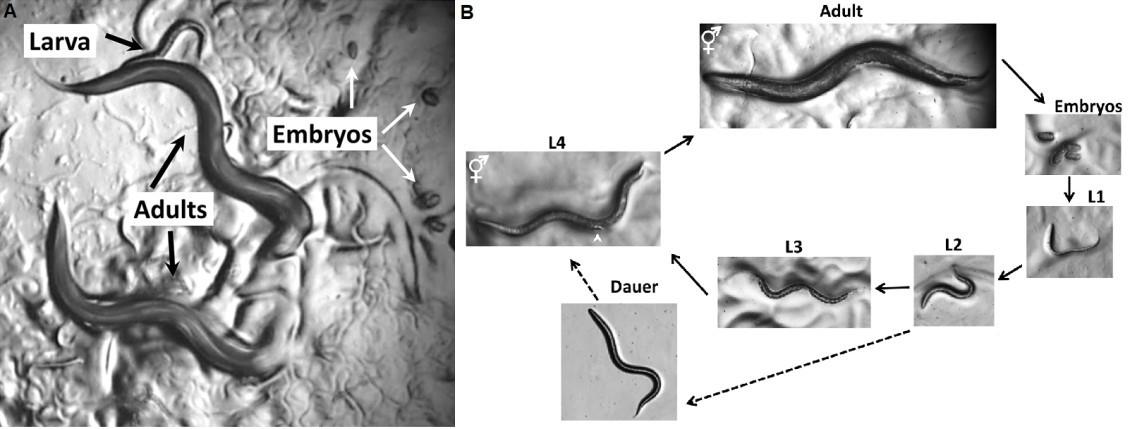
Figure 1. C. elegans life cycle from egg-laying to adult worms. A. C. elegans viewed through the dissecting microscope. B. Life Cycle of C. elegans. Animals increase in size throughout the four larval stages. Photographs were taken on Petri dishes (note the bacterial lawns in all but the dauer images). Scale bar: 0.1 mm.
DiI staining of worms
Adult worms (approximately 100 worms per condition) fed with E. coli OP50- and B. subtilis NCIB3610- strain fed adult worms are collected, washed, and resuspend in 1 mL of M9 buffer, and then mixed with 5 µL of a 1:200 dilution of DiI solution (prepared from stock solution, see Recipe 10).
Incubate on a shaker (20 × g, ~75 rpm) at room temperature for 2–3 h.
Wash worms with M9 (1–3 washes) and transfer the labeled worms onto agar pads.
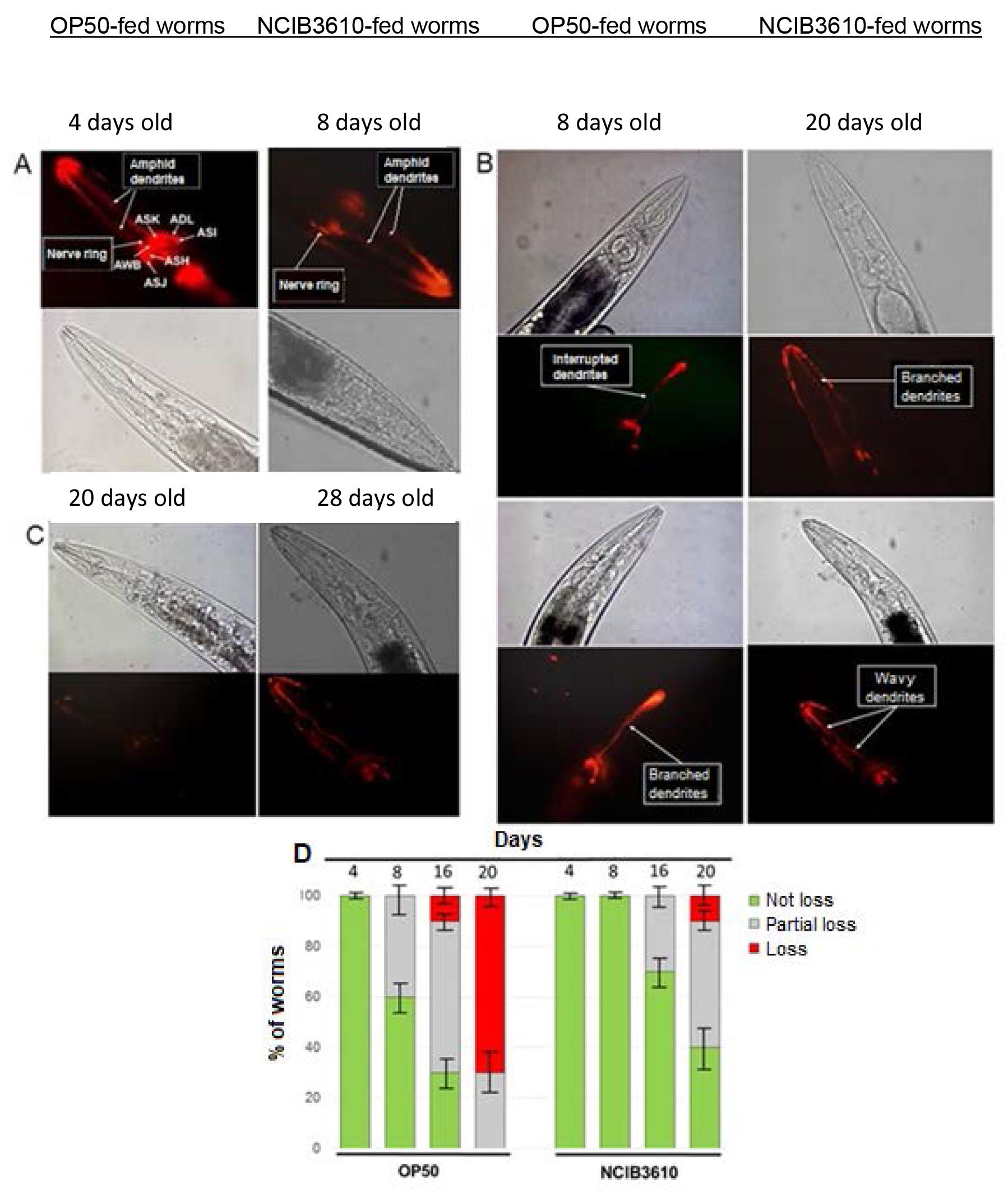
Figure 2. DiI-stained C. elegans neurons. Neuronal morphological changes of aging worms colonized by OP50 or NCIB3610 bacteria. Aging wild-type worms, colonized by OP50 or NCIB3610 bacterial cells (right and bottom rectangles for A and C; and B, respectively) at different ages, were labeled with fluorescent DiI to highlight amphid neuron morphology: A, normal morphology or no neuronal loss; B, partial neuronal alterations or partial neuronal loss; and C, total neuronal deterioration or total neuronal loss. The top and bottom micrographs (phase contrast and fluorescence microscopy, respectively) in A–C are representative of 10 independent worm images analyzed for each age. Arrows in A indicate the location of the chemosensory worm neurons (i.e., ASK, ADL, ASI, ASH, ASJ, and AWB), and arrows in B indicate some of the age-associated neuronal alterations. D. Semi-quantification of age-related neurodegeneration. Ten N2 worms colonized with OP50 or NCIB3610 bacteria, cultured on NGM plates at 20°C, were taken at the indicated times, processed, and labeled with DiI to determine the degree of age-related neuronal deterioration (not loss, partial loss, or loss). Results are expressed as a percentage of the initial population of worms (n = 100) ± S.E.M. Images from Cogliati et al. (2020) with permission.Obtain the images of the worms
Mount the labeled worms onto a 2% agar pad on a glass slide using 0.1 M sodium azide (the azide acts as an anesthetic for the worms) and enclose with a coverslip.
Use the 40× objective to visualize the labeled worms.
Neuron degeneration can be examined over time with an Olympus FV1000 laser confocal scanning microscope using the Zen program, and a semi-quantitative analysis can be performed.
Data analysis
The worms are analyzed for the absence of amphid neuron architecture (complete loss), the presence of a complete and intact set of amphid neurons (no loss), or the presence of at least one single structural abnormality, such as wavy, branched, or interrupted dendrites (partial loss) (see Figure 2). All assays are performed at least three times in duplicate. Mean survival days, standard error of the mean (S.E.M.), intervals of mean survival days with 95% confidence, and equality p values to compare averages are calculated by log-rank and Kaplan-Meier tests, using the OASIS program. The S.E.M. values are used in the figures; p < 0.05 was considered statistically significant.
Notes
When worms are old adults, instead of centrifuging, let them settle on their own, since centrifugation can damage them as they are very weak.
When mounting the worms on slices, cut off the end of the pipette tips approximately 0.5 cm to create a broader opening, which will help avoid breaking the worms while transferring them.
For a better appreciation of the staining with DiI, turn off the light when observing the worms under a microscope.
Recipes
M9 buffer
3 g KH2PO4
6 g Na2HPO4·7H2O
5 g NaCl
Water up to 1 L Autoclave, then add 1 mL of 1 M MgSO4
Nematode growth medium (NGM) agar for 1 L medium
3.0 g NaCl
20 g Bacto agar
2.5 g Bacteriological peptone
Autoclave to sterilize the agar, cool to 55°C, then add:
1 mL of 1 M MgSO4
1 mL of 1 M CaCl2
1 mL of 5 mg/mL cholesterol in absolute ethanol
25 mL of 1 M Potassium phosphate buffer (see Recipe 6)
1 M MgSO4·7H2O
Dissolve 6 g MgSO4 heptahydrate in 50 mL of dH2O
Autoclave (121°C for 20 min)
Store at room temperature
5 mg/ml cholesterol
Dissolve 0.25 g of cholesterol in 50 mL of 100% ethanol
Do not autoclave
1 M CaCl2
Dissolve 5.55 g CaCl2 dihydrate in 50 mL of dH2O
Autoclave (121°C for 20 min)
Store at room temperature
Potassium phosphate buffer (1 M KPO4, pH 6.0)
108.3 g KH2PO4
35.6 g K2HPO4
Water up to 1 L
Autoclave (121°C for 20 min)
Store at room temperature
LB media
20 g LB broth dissolve in 1 L ddH2O
Autoclave (121°C for 20 min)
Bleaching solution
1 mL of 3% hypochlorite solution
2.5 mL of 1 N NaOH
0.5 mL of dH2O
1 N NaOH
Dissolve 2 g NaOH in 50 mL of dH2O
DiI stock solution
Stock solution is 2 mg/mL in dimethyl formamide
Store at -20°C in a foil-wrapped tube
Dilute the stock solution 1:200 in M9. Some dye will precipitate when you do this.
Acknowledgments
This work was supported by CONICET (Consejo Nacional de Investigaciones Científicas y Técnicas) and FONCyT (Fondo para la Investigación Científica y Tecnológica) with the aid of the Pew Latin-American Program in Biological Sciences (Philadelphia, USA), the Fulbright Committee (Washington, DC, USA) and former Fundación Antorchas (Buenos Aires, Argentina). We modified the media and NGM plate’s preparation from Stiernagle T. Maintenance of C. elegans. WormBook. 2006 11:1-11. We modified C. elegans synchronization from Montserrat Porta-de-la-Riva et al. (2012). Basic Caenorhabditis elegans Methods: Synchronization and Observation. Journal of Visualized Experiments; 64, e4019, 1-9. The protocol is based on “Dil and diO: versatile fluorescent dyes for neuronal labelling and pathway tracing” (Honig and Hume, 1989).
Competing interests
There are no conflicts of interest or competing interests.
References
- Alexander, A., Marfil, V. and Li, C. (2014). Use of Caenorhabditis elegans as a model to study Alzheimer’s disease and other neurodegenerative diseases. Front Genet 5: 279.
- Cogliati, S., Clementi, V., Francisco, M., Crespo, C., Arganaraz, F. and Grau, R. (2020). Bacillus subtilis Delays Neurodegeneration and Behavioral Impairment in the Alzheimer's Disease Model Caenorhabditis elegans. J Alzheimers Dis 73(3): 1035-1052.
- Godement, P., Vanselow, J., Thanos, S. and Bonhoeffer, F. (1987). A study in developing visual systems with a new method of staining neurones and their processes in fixed tissue. Development 101(4): 697-713.
- Griffin, E., Caldwell, K. and Caldwell, G. (2017). Genetic and pharmacological discovery for Alzheimer’s disease using Caenorhabditis elegans. ACS Chem Neurosci 8: 2596-2606.
- Hofmann, M. H. and Bleckmann, H. (1999). Effect of temperature and calcium on transneuronal diffusion of DiI in fixed brain preparations. J Neurosci Methods 88(1): 27-31.
- Honig, M. G. and Hume, R. I. (1986). Fluorescent carbocyanine dyes allow living neurons of identified origin to be studied in long-term cultures. J Cell Biol 103(1): 171-87.
- Honig, M. G. and Hume, R. I. (1989). Dil and diO: versatile fluorescent dyes for neuronal labelling and pathway tracing. Trends Neurosci 12(9): 333-335, 340-331.
- Porta-de-la-Riva, M., Fontrodona, L., Villanueva, A. and Ceron, J. (2012). Basic Caenorhabditis elegans methods: synchronization and observation. J Vis Exp(64): e4019.
- Stiernagle, T. (2006). Maintenance of C. elegans. WormBook. 1-11. Doi: 10.1895/wormbook.1.101.1.
- Toth, M. L., Melentijevic, I., Shah, L., Bhatia, A., Lu, K., Talwar, A., Naji, H., Ibanez-Ventoso, C., Ghose, P., Jevince, A., et al. (2012). Neurite sprouting and synapse deterioration in the aging Caenorhabditis elegans nervous system. J Neurosci 32(26): 8778-8790.
- Wollenberg, A. C., Visvikis, O., Alves, A. F. and Irazoqui, J. E. (2013). Staphylococcus aureus killing assay of Caenorhabditis elegans. Bio-protocol 3(19): e916.
- Wormatlas.org (DiI and DiO Staining in C. elegans).
- Yankner, B. A., Lu, T. and Loerch, P. (2008). The aging brain. Annu Rev Pathol 3:41-66.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Crespo, C. and Grau, R. (2022). Analysis of Caenorhabditis elegans Aging-related Neurodegeneration in Chemosensory Neurons. Bio-protocol 12(14): e4473. DOI: 10.21769/BioProtoc.4473.
Category
Neuroscience > Neuroanatomy and circuitry > Animal model
Developmental Biology > Morphogenesis
Biological Sciences > Biological techniques > Microbiology techniques
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









