- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Phytophthora sojae Transformation Based on the CRISPR/Cas9 System
Published: Vol 12, Iss 6, Mar 20, 2022 DOI: 10.21769/BioProtoc.4352 Views: 3493
Reviewed by: Zhibing LaiFernando A Gonzales-ZubiateAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Gene Replacement by a Selectable Marker in the Filamentous Fungus Magnaporthe oryzae
Nalleli Garcia [...] Jessie Fernandez
Sep 5, 2023 2152 Views

CRISPR/Cas9 Ribonucleoprotein-Mediated Mutagenesis in Sporisorium reilianum
Janina Werner [...] Gunther Doehlemann
Apr 20, 2024 2558 Views

Creating Loss-of-Function Mutants of Neurospora crassa Using a Novel CRISPR/Cas9 System
Stefanie Grüttner
Jan 5, 2026 232 Views
Abstract
Phytophthora sojae is a model species for the study of plant pathogenic oomycetes. The initial research on gene function using Phytophthora was mainly based on gene silencing technology. Recently, the CRISPR/Cas9-mediated genome editing technology was successfully established in P. sojae and widely used in oomycetes. In this protocol, we describe the operating procedures for the use of CRISPR/Cas9-based genome editing technology and PEG-mediated stable transformation of P. sojae protoplasts. Two plasmids were co-transformed into P. sojae: pYF515 expressing Cas9 and the single guide RNA, and the homologous replacement vector of the candidate gene. Finally, the ORF of candidate gene were replaced with the ORF of the entire hygromycin B phosphotransferase gene (HPH), to achieve precise knockout.
Keywords: CRISPR/Cas9Background
The probability of homologous recombination of Phytophthora itself is extremely low, and it is difficult to use methods based on homologous recombination to knock out genes. Therefore, a CRISPR/Cas9-based technology was developed to allow gene editing in P. sojae (Fang and Tyler, 2016). CRISPR/Cas9 is an adaptive immune response found in Bacteria and Archaea to prevent phage infection (Deveau et al., 2010; Garneau et al., 2010; Horvath and Barrangou, 2010). The CRISPR/Cas9 system consists of two components, namely Cas9 and single guide RNA (sgRNA) (Hsu et al., 2014). The single guide RNA (sgRNA) contains 20 nucleotides, which can mediate Cas9 to target the specific sequence region based on the principle of double-strand complementarity; Cas9 protein acts as a nuclease to cut specific DNA sites to produce double-strand break nicks (DSB).
In the presence of a homologous template (donor DNA), the cell uses the homologous template to repair, resulting in the homologous replacement of the target gene by the donor DNA to complete precise gene editing. Here, we provide detailed steps for the transformation of P. sojae based on the CRISPR/Cas9 system, including sgRNA design, Cas9-sgRNA plasmid construction, homologous replacement vector construction, P. sojae transformation, and detection of mutations, using Ps139767 as an example.
Materials and Reagents
Consumables
Sterile pipette tips (10 μL, 100 μL, 1,000 μL, 5 mL)
Petri dishes (60 mm × 15 mm, 90 mm ×15 mm)
250 mL Erlenmeyer flasks
Miracloth
Stainless steel laboratory spatulas
50 mL Falcon tubes
Microcentrifuge tubes (1.5 mL, 2.0 mL)
200 μL PCR tubes
20 mL Syringe
Beaker (50 mL, 200 mL)
Bacteria filter (0.45 nm)
Parafilm
Competent cells
E. coli JM109 competent cells (homemade)
Plasmids
pBluescript SK II+(pBS-SK II+)
pYF515 (constructed by Fang and Tyler, 2016) (Figure 1)
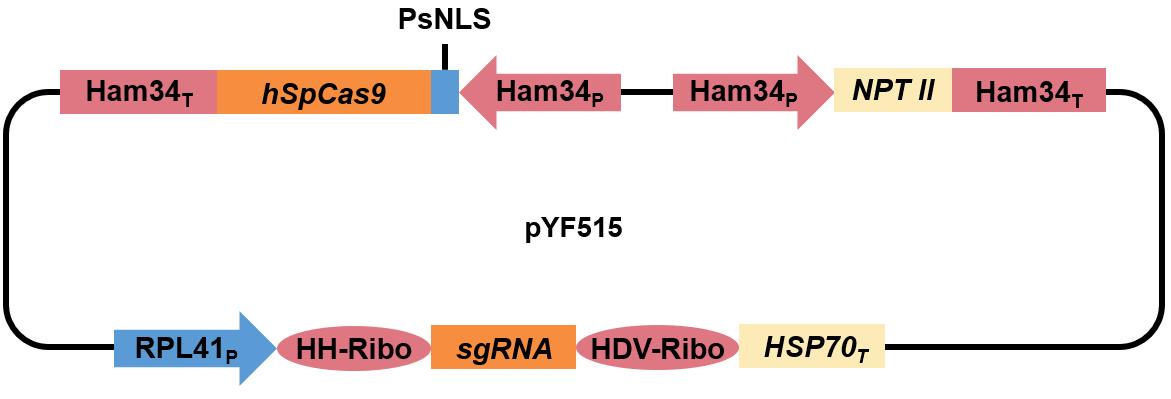
Figure 1. The pYF515 vector expresses both the Cas9 gene and the sgRNA. The expression of Cas9 and the selection marker NPT II is driven by the Ham34 promoter, and the transcription of sgRNA [including the flanking ribozyme, hammerhead ribozyme (HH ribozyme) and HDV ribozyme] is driven by the RPL41 promoter.
Primers (see Table 1)
Table 1. List of oligonucleotides (see Supplementary Information)
Primer name Sequence ( 5'→3' ) 139767-sgRNA1-F CTAGCTAGCCCGTCACTGATGAGTCCGTGAGGAC 139767-sgRNA1-R CTAGGGTCTCGAAACCAATGGTGACTACACCGTCAGACGAGCTTACTCGTTTCG 139767-sgRNA2-F CTAGCTAGCTAGTAGCTGATGAGTCCGTGAGGAC 139767-sgRNA2-R CTAGGGTCTCGAAACCCACTAGGTTGCAGTAGTAGGACGAGCTTACTCGTTTCG 139767-up1kb-F CTATAGGGCGAATTGGGTACCGCATTTCCGCAGTTCTCGTC 139767-up1kb-R GTCGCGGTGAGTTCAGGCATCGTCTACCTCCACTACGCG 139767-down1kb-F GTCCGAGGGCAAAGGAATAGACAAGCTACTCTTAGACTTTT 139767-down1kb-R AGGGAACAAAAGCTGGAGCTCAACGCAAGCACTGTCAAAGC HPH-F ATGCCTGAACTCACCGCGAC HPH-R CTATTCCTTTGCCCTCGGAC 139767-genome-F TTTGAAGACAAAAGCGGGCG 139767-genome-R TACTCGACGATACAGCACGC 139767-overup-F1 GGCCCGTGAATAAAACCCCT Hph-overup-R1 ACCATCGGCGCAGCTATTT Hph-overdown-F2 CGGGGATTCCCAATACGAGG 139767-overdown-R2 TAGCGTGTGTCAGATCCACC M13F GTAAAACGACGGCCAGT M13R CAGGAAACAGCTATGAC Strain
Phytophthora sojae strain P6497
E. coli JM109 (KangTi Life Technology, catalog number: KTSM107L)
Enzymes and buffers
BsaI and 10× Cutsmart Buffer (New England Biolabs, catalog number: R3733S)
NheI (New England Biolabs, catalog number: R3131S)
T4 DNA Ligase and 10× T4 DNA Ligase Buffer (Takara, catalog number: 2011A)
2× Phanta® Max Master Mix (Dye Plus) (Vazyme, catalog number: P525)
TaKaRa TaqTM,10× PCR Buffer(Mg2+ plus) and dNTP Mixture (Takara, catalog number: R001)
Kits
FastPure® Gel DNA Extraction Mini Kit (Vazyme, catalog number: DC301)
ClonExpress® ULtra One Step Cloning Kit (Vazyme, catalog number: C115)
FastPure® Plasmid Mini Kit (Vazyme, catalog number: DC201-01)
DNAsecure New Plant Genomic DNA Extraction Kit (TIANGEN BIOTECH, catalog number: DP320)
TaKaRa MiniBEST DNA Fragment Purification Kit Ver.4.0 (Takara, catalog number: 9761)
Antibiotics
Ampicillin (to a final concentration of 50 mg/mL) (Aladdin, catalog number: 69-52-3)
G418 (to a final concentration of 50 mg/mL) (Warbio, 108321-42-2)
Media (see Recipes)
Luria-Bertani agar plates + 50 mg/mL ampicillin
Luria-Bertani liquid medium
10% V8 agar plates +50 mg/mL G418
10% V8 liquid medium
Nutrient pea broth and agar medium (agar plates and liquid)
Mannitol-pea broth (agar plates and liquid)
Reagents
V8 vegetable juice (Campbell Soup Company, catalog number: 051000153159)
Frozen green peas
Lysing Enzymes (Sigma-Aldrich, catalog number: L1412)
CELLULYSIN Cellulase (Millipore, catalog number: 9012-54-8)
Bacto tryptone (OXOID, catalog number: 73049-73-7)
Yeast extract (Sigma-Aldrich, catalog number: 8013-01-2)
NaCl (Aladdin, catalog number: 7647-14-5)
Agar (Aladdin, catalog number: 9002-18-0)
CaCO3 (Sangon, catalog number: 471-34-1)
CaCl2 (Sangon, catalog number: 10043-52-4)
KCl (Sangon,catalog number: 7447-40-7)
PEG4000 (Aladdin, catalog number:25322-68-3)
Biotin (Aladdin, catalog number:58-85-5)
Folic acid (Aladdin, catalog number: 59-30-3)
L-inositol (Sigma-Aldrich, catalog number: 551-72-4)
Nicotinic acid (Aladdin, catalog number: 59-67-6)
Pyridoxine-HCl (Aladdin, catalog number: 58-56-0)
Riboflavin (Aladdin, catalog number: 83-88-5)
Thiamine-HCl (Aladdin, catalog number: 59-43-8)
FeC6H5O7·3H2O (Sangon, catalog number: 17217-76-4)
ZnSO4·7H2O (Sangon, catalog number: 7446-20-0)
CuSO4·5H2O (Sangon, catalog number: 7758-99-8)
MgSO4·H2O (Sangon, catalog number: 14567-64-7)
H3BO3 (Phytotech, catalog number: 10043-35-3)
MoO3 (Phytotech, catalog number: 1313-27-5)
KH2PO4 (Sangon, catalog number: 7778-77-0)
K2HPO4 (Sangon, catalog number: 7758-11-4)
KNO3 (Aladdin, catalog number: 7757-79-1)
D-sorbitol (Aladdin, catalog number: 50-70-4)
D-mannitol (Aladdin, catalog number: 69-65-8)
Glucose (Aladdin, catalog number: 50-99-7)
MgCl2·6H2O (Diamond, catalog number: 7791-18-6)
MES (Aladdin, catalog number: 4432-31-9)
Tris-HCl (Diamond, catalog number: 1185-53-1)
EDTA (Phytotech, catalog number: 60-00-4)
Hexadecyl trimethyl ammonium Bromide(CTAB) (Phytotech, catalog number: 57-09-0)
Solutions
0.8 M Mannitol solution, autoclave at 121°C for 20 min
0.5 M CaCl2 solution, sterilize using a 0.45-mm filter
0.5 M KCl solution, sterilize using a 0.45-mm filter
Luria-Bertani medium (see Recipes)
10% V8 agar (see Recipes)
Pea broth (see Recipes)
Nutrient pea broth and agar medium (see Recipes)
0.5 M MES-KOH solution (see Recipes)
W5 solution (see Recipes)
MMg solution (see Recipes)
Enzyme solution (see Recipes)
40% PEG solution (see Recipes)
Mannitol-pea broth (see Recipes)
2% CTAB solution (see Recipes)
Vitamin stock (see Recipes)
Trace elements (see Recipes)
Equipment
Pipettes (Eppendorf)
Rotating mixer (Select BioProducts, model: SBS550-2)
PCR machine (Bio-Rad, model: T100TM)
Light microscope (Leica, model: DM500)
Incubator (25°C) (Ningbo Jiangnan, model: HWS-1000)
Water bath (42°C) (SHELLAB, model: SWB7-2)
Tabletop centrifuge(4°C) (Thermofisher, model: ST8)
Incubator-shaker (37°C) (Crystal, model: IS-RSD3)
DNA electrophoresis apparatus (Bio-Rad, model: 1704469)
Software
sgRNA design software: EuPaGDT (http://grna.ctegd.uga.edu/)
Sequence alignment: SeqHunter2 (https://sourceforge.net/projects/seqhunter2)
Procedure
Screening and analysis of sgRNA targets
Use EuPaGDT to find all qualified sgRNAs in the target gene sequence.
Enter the gene ID in the "Job Name" column, select the "SpCas9" option in the "RNA guided nuclease selection" column, select P. sojae P6497 from Oomycetes in the "Genome (expand a category and choose one)" column, enter the gene sequence in the "Sequence" column, and click "get guide RNA" to obtain all the potential sgRNA targets in the target sequence.
Off-target analysis of candidate sgRNA.
Find an sgRNA target in the sense strand and antisense strand of the candidate gene, select the core region of candidate sgRNA (the 12 bp seed sequence at the 3'end and the NGG PAM sequence), and compare it with the genome sequence of P. sojae, using sequence alignment tools in Seqhunter2 or online webserver EumicrobeDB (http://www.eumicrobedb.org). Choose sgRNA targets with good specificity.
Construction of CRISPR/Cas9 plasmid
sgRNA synthesis (taking Ps139767 as an example)
Primer design
The sequence of HH-RIbo +sgRNA+HDV-Ribo is as follows:
NNNNNNCTGATGAGTCCGTGAGGACGAAACGAGTAAGCTCGTCNNNNNN(N)14GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTGGCCGGCATGGTCCCAGCCTCCTCGCTGGCGCCGGCTGGGCAACATGCTTCGGCATGGCGAATGGGAC
(Taking 20 bp sequence TGACGGTGTAGTCACCATTG as an example)
Target fragment sequence:
5’CTAGCTAGCCCGTCACTGATGAGTCCGTGAGGACGAAACGAGTAAGCTCGTCTGACGGTGTAGTCACCATTGGTTTCGAGACCCTAG 3’
NheI:
5′… GCTAGC …3′
3′… CGATCG … 5′
BsaI:
5′… GGTCTC (N)1 …3′
3′… CCAGAG (N)5 … 5′
The primers designed are:
F: 5′ CTAGCTAGCCCGTCACTGATGAGTCCGTGAGGAC 3′
R: 5′ CTAGGGTCTCGAAACCAATGGTGACTACACCGTCAGACGAGCTTACTCGTTTCG 3′
Clone sgRNA
The sgRNA vector template was pYF515 containing sgRNA fragment.
Setup reactions for PCR amplification using:
2× Phanta® Max Master Mix (Dye Plus) 25 μL Forward oligo (10 µM) 1 μL Reverse oligo (10 µM) 1 μL The template 1 μL Add ddH2O to 50 μL Amplify the targeted region by PCR [The annealing temperature (x) depends on the primers]:
95°C 5 min 1 cycle 95°C 30 s 32 cycles x°C 30 s 72°C 10 s 72°C 5 min 1 cycle 4°C hold 1 cycle
Enzyme digestion of sgRNA fragments.
BsaI 0.5 μL NheI 0.5 μL 10× Cutsmart Buffer 5 μL sgRNA fragment 20 μL Add ddH2O to 50 μL Enzymatic digestion of pYF515 vector.
BsaI 4 μL NheI 4 μL 10× Cutsmart Buffer 20 μL The template 8 μg Add ddH2O to 200 μL Use the MiniBEST DNA Fragment Purification Kit to purify the digested vector and fragment respectively.
Connect the sgRNA fragment to the pYF515 vector.
T4 DNA Ligase 1 μL 10× T4 DNA Ligase Buffer 1 μL Linearization vector 1–2 μL sgRNA fragment 7 μL Place it at 10–16°C for 2 h.
Use 10 μL of the ligation product to transform E. coli JM109 competent cells (homemade).
Use the FastPure Plasmid Mini Kit to extract the plasmids and send them for sequencing with M13F and M13R primers.
Construction of the replacement vector
Amplify the sequence 1 kb upstream and 1 kb downstream of the target gene, and the replacement fragment HPH.
After cutting the gel, use the FastPure® Gel DNA Extraction Mini Kit to recover the fragments.
Multi-fragment connection.
Optimal cloning vector usage = [0.02× number of base pairs of cloning vector] ng (0.03 pmol)
Optimal usage amount of each fragment = [0.02× number of base pairs per fragment] ng (0.03 pmol)
The volume (X, Y1, Y2, and Y3) of the vector and fragments depends on the number of base pairs and their concentration.
2× ClonExpress Mix 5 μL pBluescript SK II+ vector, Linearized X μL UP-1Kb fragment Y1 μL Down-1Kb fragment Y2 μL Replacement fragment Y3 μL ddH2O to 10 μL Set in the PCR machine at 50°C for 15 min; drop directly to 4°C.
Use 10 μL of the ligation product to transform E. coli JM109 competent cells (homemade).
Use the FastPure Plasmid Mini Kit to extract the plasmids and send them for sequencing with M13F and M13R primers.
PEG-mediated stable transformation of P. sojae protoplasts
Activate P. sojae on a 70 mm Nutrient pea broth and agar medium (NPB) solid plate, and cultivate it in the dark at 25°C for 3 to 4 days (Figure 2A).
Use a scalpel to cut a 3 × 3 mm fresh mycelium block from the edge of the colony, and add it into a 250 mL Erlenmeyer flask containing 50 mL of liquid NPB. Each Erlenmeyer flask can hold eight mycelium blocks. Incubate the flasks at 25°C in the dark for 2.5 to 3 days, shaking them once every half a day during the culture period. Three flasks of P. sojae culture are optimal for six DNA samples (Figure 2B).
Turn on the ultra-clean workbench and sterilize with an UV lamp for 30 min. During this period, weigh the amount of enzyme needed to prepare the enzyme solution, and the PEG needed for 40% PEG into a sterile 50 mL beaker. The tools used in the weighing process must be as clean and sterile as possible.
Filter the hyphae of P. sojae that have been cultured for 2.5 to 3 days with autoclaved gauze tied to a 200 mL beaker, place the hyphae mass into a 50 mL Falcon tube, and rinse once with about 40 mL of 0.8 M mannitol. Collect the hyphae again with another sterile gauze wrapped over a 200 mL beaker, place the hyphae in a 50 mL Falcon tube, and pour 35 mL of 0.8 M mannitol buffer over them. To sufficiently treat the hyphae with mannitol, turn the Falcon tube upside down to fully separate the hyphae. Shake and wash the hyphae with a rotating mixer at 20 rpm (0.003 × g) and at room temperature (RT) for 10 min.
During this period, prepare the enzyme solution. Add the specified volume of each component to a sterilized 50 mL beaker, stir with a pipette tip to fully dissolve, and filter into a 50 mL Falcon tube with a 0.45 nm bacterial filter.
After the hyphae have been rinsed for 10 min, collect an appropriate amount of hyphae, and add them to the Falcon tube containing the enzyme solution. To fully digest the hyphae, invert the centrifuge tube upside down, and perform enzymatic hydrolysis by centrifuging at 60 rpm and 25°C for 40–50 min (Figure 2C).
To prepare a 40% PEG solution, add the specified volume of each component, stir with a pipette tip until they are completely dissolved, filter the solution with a 0.45 nm bacterial filter into a 50 mL sterile beaker, and place it on ice for later use.
Use a 50 mL beaker with two layers of mira-cloth to filter the hyphae and collect the protoplasts. Use a 50 mL round bottom centrifuge tube to place the collected protoplasts in a horizontal centrifuge at at 240 × g and 4°C for 4 min.
Note: All subsequent operations require the protoplasts to be kept at low temperature (Figure 2D).
Discard the supernatant, add approximately 10 mL of pre-cooled W5 solution to gently resuspend the protoplasts, then add W5 solution to 35 mL, and centrifuge at 240 × g and 4°C for 4 min.
Discard the supernatant and add approximately 10 mL of pre-cooled W5 solution to gently resuspend the protoplasts. Place on ice for 30 min, then centrifuge it at 240 × g and 4°C for 4 min.
Discard the supernatant, add pre-chilled MMg solution according to the number of samples to resuspend the protoplasts, to achieve a protoplast concentration of 2 × 106/mL. Keep at RT for 10 min.
Add at least 30 μg of transformed plasmids to the bottom of several new 50 mL Falcon tubes placed on ice.
Add 1 mL of protoplasts to each Falcon tube, mix completely by gently tapping the tube, and keep the samples on ice for 5–10 min.
Add 580 μL of PEG solution to each Falcon tube along the tube wall three times, for a total of 1.74 mL of PEG solution. Gently rotate the Falcon tube during the addition, so that the PEG can flow into the mixture of plasmids and protoplasts along the wall of the tube. After adding the total amount of PEG solution, gently tap the tube to mix well, and place on ice for 20 min (Figure 2E).
Add the storage solution of ampicillin (50 mg/mL) to mannitol-pea broth (PM) medium to make the final concentration of ampicillin 50 μg/mL, mix it well, and set aside for later use.
Add 2 mL of PM medium to each Falcon tube, slowly invert it once, and place on ice for 2 min.
Add 8 mL of PM medium to each Falcon tube, slowly invert it once, and place on ice for 2 min.
Add 10 mL of PM medium to each Falcon tube. Incubate it in a tilted stand at 25°C, and regenerate overnight (Figure 2F).
Take 5 μL of the culture that has been regenerated overnight (~14 h) and check the regeneration under a microscope. Centrifuge the Falcon tube at 430 × g for 5 min to precipitate the regenerated hyphae (Figure 2G).
Discard the supernatant to leave about 5 mL of liquid in the tube, gently pipette to suspend it, and then add 35 mL of pea/0.5 M mannitol agar medium containing 25 μg/mL G418. Mix up and down, pour it into a Petri dish, blow-dry the water vapor, and incubate at 25°C in the dark (Figure 2H).
After 2–3 days, after new hyphae grow out, cover with 15 mL of V8 agar medium containing 50 μg/mL G418, blow-dry the water vapor, and continue culturing at 25°C in the dark (Figure 2I).
After approximately 3 to 4 days, the mycelium will grow again. Cut a single regenerated small colony with dense hyphae and transfer them to a V8 plate, containing 50 μg/mL G418 for culture. The regenerated strains obtained are initially considered to be knockout mutant, used for subsequent identification, and phenotypic analysis.

Figure 2. PEG-mediated transformation procedure of P. sojae. (A) A colony of P. sojae (strain P6497) was grown on nutrient pea broth for 3–5 days. (B) An agar plug was used to inoculate nutrient pea broth in a flask. The image was taken after 3 days of inoculation. (C) P. sojae mycelium digestion using a flip mixer. (D) Filter the hyphae and collect the protoplasts in a 50 mL beaker with two layers of mira-cloth. (E) Slowly add 40% PEG solution along the wall of the Falcon tube and let the droplet slide to the bottom of the tube. (F) Incubate protoplasts at 25°C in a tilted and static manner, and allow them to regenerate overnight. (G) Check the regenerated hyphae under a microscope. (H) Pour plates with regenerated protoplasts. (I) After new hyphae grow out, cover with V8 agar medium containing 50 μg/mL G418.
Screening and verification of knockout mutants
It is recommended that the resistant transformants obtained by Phytophthora transformation grow on G418-containing plates for 7 days, before screening transformants. Extract the genes of the transformants, then screen and verify the knockout mutants by genomic PCR and sequencing methods.
CTAB method for crude extraction of mycelial genomic DNA:
Scrape an appropriate amount of mycelium into a 2 mL centrifuge tube, then add 500 μL of 2% CTAB and 500 μL of chloroform, and treat it in a shaker at 37°C for 1–2 h.
Centrifuge the tube at 16,000 × g for 10 min. Take 400 μL of the supernatant, transfer it to a new 1.5 mL centrifuge tube, add 800 μL of absolute ethanol, mix well, and place it at -20°C for precipitation for more than 30 min.
Centrifuge the tube at 16,000 × g for 10 min, discard the supernatant, centrifuge again for 2 min, and use a pipette to completely aspirate the supernatant.
Place the centrifuge tube in an oven at 50°C for 10–20 min, to allow the ethanol to volatilize.
Dissolve with 30 μL of ddH2O and store at -20°C.
Alternative: Refined extraction of genomic DNA
Use Tiangen new plant genome extraction kit (DP320-50), and carry out the operation in accordance with the manufacturer’s instructions.
Screening for knockout mutants
Use the Ps139767 gene internal primer 139767-genome-F/R to conduct a PCR, to verify whether the Ps139767 sequence exists in the genome. Select the transformants which contain no Ps139767 signal as candidate knockout mutants.
Perform the PCR with primer 139767-overup-F1 outside the upper arm 1 kb with HPH internal primer Hph-overup-R1, and HPH internal primer Hph-overdown-F2 with primer 139767-overdown-R2 outside the lower arm 1kb, to verify that the replacement fragment was successfully integrated into the correct position in the genome (Figure 3).
Further verify the PCR results by sequencing with M13F and M13R, to screen the candidate knockout mutants.
Set up the reaction for PCR amplification of genomic DNA extracted from candidate mutants:
dNTP 2 μL 10× PCR Buffer (Mg2+ plus) 2.5 μL rTaq 0.25 μL Forward oligo (10 μM) 0.25 μL Reverse oligo (10 μM) 0.25 μL The template 1.5 μL Add ddH2O to 25 μL Amplify the targeted region by PCR [The annealing temperature (x) and extension time (n) depend on the primers]:
95°C 5 min 1 cycle 95°C 30 s 32 cycles x°C 30 s 72°C n min 72°C 5 min 1 cycle 4°C hold 1 cycle
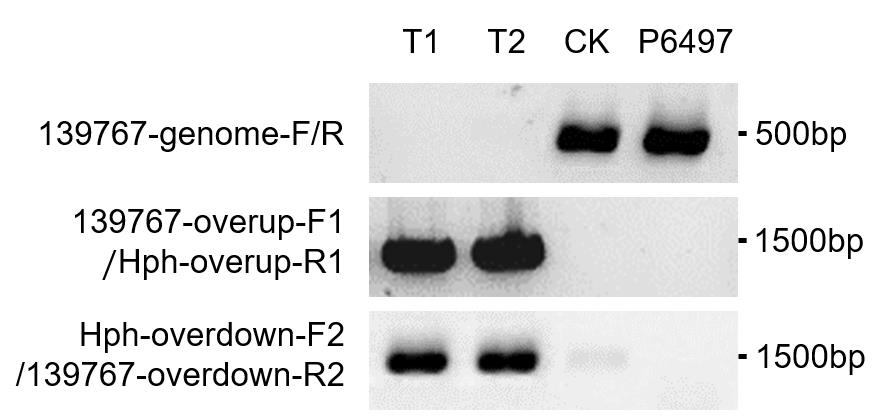
Figure 3. Analysis of genomic DNA from Ps139767 candidate transformants.
Recipes
Luria-Bertani medium (1 L)
10 g Bacto tryptone
5 g Yeast extract
10 g NaCl
1.5% agar (w/v, for solid medium)
Autoclave and store at room temperature before use.
10% V8 agar (1 L)
100 mL of V8 juice
900 mL of ddH2O
1 g CaCO3
1.5% agar (w/v, for solid medium)
Autoclave and store at room temperature before use.
Pea broth (1 L)
120 g Frozen peas
700 mL of ddH2O
Autoclave and collect supernatant
Vitamin stock (300 mL)
10 μL of 0.02 g/mL Biotin
10 μL of 0.02 g/mL Folic acid
0.012 g of L-inositol
0.06 g Nicotinic acid
0.18 g Pyridoxine–HCl
0.015 g Riboflavin
0.38 g Thiamine-HCl
Add ddH2O to 300 mL
Filter-sterilize and then store (up to 1 year) at 4°C.
Trace elements (400 mL)
0.215 g FeC6H5O7·3H2O
0.15 g ZnSO4·7H2O
0.03 g CuSO4·5H2O
0.015 g MgSO4·H2O
0.01 g H3BO3
0.007g MoO3
Add ddH2O to 400 mL
Filter-sterilize and then store at 4°C (up to 1 year).
Nutrient pea broth and agar medium (1 L)
1.0 g KH2PO4
1.0 g K2HPO4
3.0 g KNO3
0.5 g MgSO4
0.1 g CaCl2
2.0 g CaCO3
5.0 g D-sorbitol
5.0 g D-mannitol
5.0 g Glucose
2.0 g Yeast extract
2.0 mL of Vitamin stock (see recipe above)
2.0 mL of Trace elements (see recipe above)
900 mL of Pea broth (see recipe above)
Adjust to 1 L with ddH2O
1% agar (w/v, for solid medium)
Autoclave at 121°C for 20 min and store at 4°C.
Enzyme solution (20 mL)
10 mL of 0.8 M mannitol
0.8 mL of 0.5 M KCl
0.8 mL of 0.5 M MES
0.4 mL of 0.5 M CaCl2
0.15 g Lysing enzymes from Trichoderma harzianum
0.06 g CELLULYSIN cellulase
8 mL of ddH2O
W5 solution (250 mL)
0.093 g KCl
4.6 g CaCl2·2H2O
2.25 g NaCl
7.8 g Glucose
Add ddH2O to 250 mL
Autoclave at 121°C for 20 min and store at 4°C.
0.5 M MES-KOH solution
4.88 g MES
40 mL of ddH2O
Adjust pH to 5.7 using 1 M KOH and filter sterilization.
MMg solution (250 mL)
18.22 g Mannitol
0.76 g MgCl2·6H2O
2 mL of 0.5 M MES (pH = 5.7)
Add ddH2O to 250 mL
Autoclave at 121°C for 20 min and store at 4°C.
40% PEG solution (w/v, 31.5 mL, enough for 12 samples)
12 g PEG4000
7.5 mL of 0.8 M Mannitol
6 mL of ddH2O
6 mL of 0.5 M CaCl2
Filter-sterilize and store at 4°C.
Mannitol-pea broth (1 L)
91.1 g Mannitol
1.32 g CaCl2·2H2O
2 g CaCO3
900 mL of pea broth (see recipe above)
Add ddH2O to 1 L
1% bacto agar (w/v, for solid medium)
2% CTAB solution (800 mL)
80 mL of 1 M Tris-HCl
32 mL of 0.5 M EDTA
65.52 g NaCl
16 g CTAB
Add ddH2O to 800 mL
Dissolve in 65°C water bath and adjust pH to 8.0 using 1 M NaOH.
Acknowledgments
We thank Dr Brett Tyler (Oregon State University, Corvallis) for providing us with pYF515 and pBluescript SK II+ plasmids. This work was supported by grants to W. Y. from Chinese Modern Agricultural Industry Technology System (CARS-004-PS14), grants to Y. W. from China National Funds for Innovative Research Groups (31721004), grants to Min Qiu from the China Postdoctoral Science Foundation (2020M681642).
Competing interests
There are no conflicts of interest or competing interests.
References
- Deveau, H., Garneau, J. E. and Moineau, S. (2010). CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol 64: 475-493.
- Fang, Y. and Tyler, B. M. (2016). Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol Plant Pathol 17(1): 127-139.
- Garneau, J. E., Dupuis, M. E., Villion, M., Romero, D. A., Barrangou, R., Boyaval, P., Fremaux, C., Horvath, P., Magadan, A. H. and Moineau, S. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468(7320): 67-71.
- Horvath, P. and Barrangou, R. (2010). CRISPR/Cas, the immune system of bacteria and archaea. Science 327(5962): 167-170.
- Hsu, P. D., Lander, E. S. and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157(6): 1262-1278.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Cao, J., Qiu, M., Ye, W. and Wang, Y. (2022). Phytophthora sojae Transformation Based on the CRISPR/Cas9 System. Bio-protocol 12(6): e4352. DOI: 10.21769/BioProtoc.4352.
Category
Microbiology > Microbial genetics > CRISPR-Cas9
Molecular Biology > DNA > Transformation
Molecular Biology > DNA > Mutagenesis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









