- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Rapid Protocol for Direct Isolation of Osteoclast Lineage Cells from Mouse Bone Marrow
(*contributed equally to this work) Published: Vol 12, Iss 5, Mar 5, 2022 DOI: 10.21769/BioProtoc.4338 Views: 4356
Reviewed by: Salma MerchantKuo-Ching "KC" MeiYogita Jethmalani

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Isolation of CD31+ Bone Marrow Endothelial Cells (BMECs) from Mice
Alhaji Osman Smith [...] Lingyu Zeng
Nov 20, 2021 4068 Views
Primary Mouse Invariant Natural Killer T (iNKT) Cell Purification and Transduction
Gloria Delfanti [...] Giulia Casorati
Jul 5, 2023 2169 Views
Isolation and Culture of Primary Pericytes from Mouse
Tamara McErlain [...] Meera Murgai
Apr 20, 2025 2978 Views
Abstract
Osteoclast lineage cells (OLCs), including osteoclast precursors (OCPs) and mature osteoclasts (MOCs), participate in bone remodeling and mediate pathologic bone loss. Thus, it is essential to obtain OLCs for exploring their molecular features in both physiological and pathological conditions in vivo. However, the conventional protocols for obtaining OLCs ex vivo are not only time-consuming, but also unable to capture the cellular status of OLCs in vivo. In addition, the current antibody-based isolation approaches, such as fluorescence-/ magnetic-activated cell sorting, are not able to obtain pure osteoclasts because no unique surface antigen for osteoclasts has been identified. Here, we develop a rapid protocol for directly isolating OLCs from mouse bone marrow through magnetic-activated cell sorting (MACS). This protocol can rapidly enrich OCPs and MOCs, respectively, depending on the expression of the distinctive surface markers at their differentiation stages. It is optimized to isolate OLCs from four mice concurrently, of which sorting procedure could be completed within ~5 h.
Keywords: Magnetic-activated cell sortingBackground
Osteoclast lineage cells (OLCs) derived from bone marrow monocytes/macrophages (BMMs) govern bone resorption to regulate bone remodeling (Quinn et al., 1998). BMMs first differentiate into the committed osteoclast precursors (OCPs), which further fuse together to form mature osteoclasts (MOCs) (Asagiri and Takayanagi, 2007). Mounting evidence demonstrates that the dysregulation of osteoclasts could cause skeletal disorders, such as osteoporosis and Paget’s disease (Feng and McDonald, 2011; Novack and Mbalaviele, 2016). The excessive osteoclast activity and aberrant osteoclastic resorption in subchondral bone could also contribute to the progression of osteoarthritis (Zhen et al., 2013; Zhu et al., 2019; Liu et al., 2021). Thus, it is essential to directly isolate live OLCs in vivo, for studying their biological features and pathologic changes.
The laboratory mouse is a commonly used animal model for investigating osteoclast function (Elefteriou and Yang, 2011). The conventional protocol for obtaining osteoclasts relies on stimulating the primary mouse BMMs from bone marrow cells (BMCs) with macrophage colony stimulating factor (M-CSF) and receptor activator of NFκB-ligand (RANKL) in vitro (Glantschnig et al., 2003; Marino et al., 2014). This protocol requires more than one week to form the MOCs in vitro, which could not capture their real-time status in vivo. Alternatively, fluorescence-activated cell sorting (FACS) has been previously employed to directly isolate the live OLCs from BMCs in vivo (Atkins et al., 2006; Zhang et al., 2012; Madel et al., 2018). Nevertheless, the disadvantages of FACS, including time-consuming procedures, low yield, and high cost, significantly limit its widespread use (Lee and Lufkin, 2012; Almeida et al., 2014). Therefore, it is necessary to introduce more efficient isolation techniques for directly obtaining live OLCs in vivo.
Magnetic-activated cell sorting (MACS) is another frequently used technique for sorting live cells in vivo, which enables rapid cell isolation with high yield and low cost (Lee and Lufkin, 2012; Almeida et al., 2014). This technique has been previously applied to recognize single surface markers of osteoclasts, for isolating OLCs from chicken or mouse BMCs in vivo (Collin-Osdoby and Osdoby, 2012; Liu et al., 2015). Since no specific surface marker for defining OCPs or MOCs has been found until now, it is not feasible to directly distinguish OCPs and MOCs from OLCs through one single surface marker. Calcitonin receptor (CTR), which is highly expressed on osteoclasts, is commonly used for characterizing MOCs, while osteoclast-associated receptor (OSCAR) was considered a typical cell surface marker for OLCs. We have previously employed MACS to harvest the OSCAR+ OLCs from mouse bone marrow in vivo for downstream gene and miRNA expression assay (Liu et al., 2015; Li et al., 2016). Considering the distinctive expression of OSCAR and CTR throughout the differentiation stages of OLCs, OSCAR may serve as an ideal surface marker for identifying OLCs among BMCs, while CTR could be used to distinguish the MOCs (OSCAR+CTR+) from the OCPs (OSCAR+CTR–) within the OSCAR+ OLCs. Therefore, we further developed a two-step MACS method for distinguishing OCPs and MOCs from OLCs in BMCs, which demonstrated high efficiency in the direct capture of OCPs and MOCs from mice that had undergone surgery-induced osteoarthritis in our most recent study (Liu et al., 2021).
Here, we described this novel two-step MACS protocol for rapidly isolating OCPs and MOCs directly from mouse bone marrow with high yield and purity. This protocol consists of three main steps: BMC preparation, antibody-microbead labelling and magnetic sorting, and osteoclast characterization (Figure 1). In brief, the CTR+ and CTR– cell fractions were first separated from mouse BMCs by magnetic positive and negative selection. Thereafter, the OSCAR+CTR+ MOCs, among the CTR+ cell fractions, and the OSCAR+CTR– OCPs, among the CTR– cell fractions, were enriched by another magnetic positive selection. We employed this protocol to rapidly isolate the MOCs and OCPs from both ovariectomized (OVX) and sham-operated (Sham) mice. The sorting procedures can be carried out in four mice concurrently within ~5 h.

Figure 1. Schematic diagram of isolation for osteoclast lineage cells (OLCs) from mouse bone marrow
Materials and Reagents
1.5 mL microtubes (Biopioneer, catalog number: CNT-1.5F)
15 mL Falcon tubes (Corning, catalog number: 430791)
50 mL Falcon tubes (Corning, catalog number: 352070)
MS columns (Miltenyi Biotech, catalog number: 130-042-201, column capacity: up to 107 magnetically labeled cells from up to 2 × 108 total cells)
LS columns (Miltenyi Biotech, catalog number: 130-042-041, column capacity: up to 108 magnetically labeled cells from up to 2 × 109 total cells)
0.2-μm pore sized filter, bottle top (Fisher Scientific, catalog number: 595-4520)
1 mL syringe with 25G × 25 mm needle (Terumo)
100 mm Culture dish (Corning, catalog number: 354600)
30 μm pre-separation filters (Miltenyi Biotech, catalog number: 130-041-407,)
Cell strainer (BD Falcon, 40 μm, catalog number: 352340)
75% (vol/vol) Ethanol
CAUTION! Flammable. Keep away from flame and sources of ignition. Store in dry and well-ventilated areas.
QuadroMACS Starting Kit (Miltenyi Biotech, catalog number: 130-091-051)
Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher, catalog number: 11965)
Penicillin/streptomycin (Thermo Fisher, catalog number: 15140122)
Fetal bovine serum (FBS) (Thermo Fisher, catalog number: 16170-078)
Bovine serum albumin solution (BSA) (MP Biomedicals, catalog number: 810531)
Sodium chloride (NaCl) (Fisher Scientific, catalog number: 7647-14-5)
Potassium chloride (KCl) (Fisher Scientific, catalog number: 7447-40-7)
Sodium phosphate (Na2HPO4) (Fisher Scientific, catalog number: 7778-77-0)
Potassium bicarbonate (KHCO3) (Fisher Scientific, catalog number: 298-14-6)
10× red blood cell (RBC) lysis solution (Miltenyi Biotech, catalog number: 130-094-183)
Dead Cell Removal Kit (Miltenyi Biotech, catalog number: 130-090-101), including the dead cell removal MicroBeads and 20× dead cell binding (DCB) buffer stock solution
Ethylenediaminetetraacetic acid (EDTA) (Sigma, catalog number: E6758-100G)
Paraformaldehyde (Sigma-Aldrich, catalog number: 158127)
Methanol (Sigma-Aldrich, catalog number: 34860)
Triton-X100 (Sigma-Aldrich, catalog number: X100-500ML)
Toluidine Blue (Sigma-Aldrich, catalog number: 198161)
OSCAR antibody (R&D Systems, catalog number: MAB1633, Isotype: monoclonal rat IgG2A)
Biotin-conjugated CTR antibody (LifeSpan BioSciences, catalog number: LS-C523811, Isotype: Biotin-conjugated polyclonal rabbit IgG)
Anti-Rat IgG MicroBeads (Miltenyi Biotech, catalog number: 130-048-502)
Anti-Biotin Multisort Kit (Miltenyi Biotech, catalog number: 130-091-256), including the Anti-Biotin Multisort MicroBeads, MultiSort Release Reagent, and MultiSort Stop Reagent
Monoclonal Rat IgG2A Isotype Control (R&D Systems, catalog number: MAB006)
Biotin-conjugated polyclonal rabbit IgG Isotype Control (LifeSpan BioSciences, catalog number: LSC351735)
Alexa Fluor 488 goat anti-rat IgG (H+L) Cross-Adsorbed secondary antibody (Thermo Scientific, catalog number: A11006)
PE goat anti-rabbit IgG (H+L) Cross-Adsorbed secondary antibody (Thermo Scientific, catalog number: P-2771MP)
DPBS (see Recipes)
MACS buffer (see Recipes)
Complete DMEM Medium (see Recipes)
1× RBC Lysis Solution (see Recipes)
1× DCB buffer (see Recipes)
1% Toluidine Blue solution (see Recipes)
Equipment
Dissection tools (surgical scissors, microforceps)
4°C Swinging-bucket centrifuge (e.g., Allegra X-15 R, Beckman Coulter)
Hemocytometer/counting chamber (e.g., Hausser Scientific Partnership, catalog number: 3200)
FACS (e.g., BD FACS, Aria III Cell-Sorting System, BD Biosciences)
Procedure
Preparing of bone marrow cells ● TIMING 40 min
Euthanize a 3-month-old mouse by cervical dislocation under general anesthesia or other approved methods. CAUTION! All animal procedures must be carried out in accordance with your local animal ethics and experimental safety committees.
Sterilize the mouse with 70% ethanol, dissect the hind limbs, and remove the soft tissues by scraping the diaphysis off the bone. Store the bones in cold complete DMEM on ice.
Harvest the BMCs in a hood using sterilized equipment. Cut the ends of the tibia and femur just below the end of the marrow cavity. Remove the bone growth plate and insert a 27G needle attached to a 1 mL syringe containing complete medium into the spongy bone. Flush the bone marrow plug out of the shaft with complete medium repeatedly, and collect the plug into a 40 μm basket filter placed in a dish on ice. CRITICAL STEP The flushing can be stopped when the bone marrow cavity is no longer red.
Use the end of a new syringe to disperse the marrow plug through the basket filter for removing fat, clustered debris, and other unwanted materials.
Pass the cell-containing medium through cell filters into a 15 mL conical tube to obtain a single cell suspension. Centrifuge at 300 × g and 4°C for 10 min to pellet the cells, and then discard the supernatant.
Add 10 mL of 1× RBC Lysis Solution into the BMCs from one mouse. Gently vortex for 5 s and incubate for 10 min at room temperature. CRITICAL STEP Avoid the high-speed vortex (Recommended speed ≤ 600 RPM).
Centrifuge at 300 × g and 4°C for 5 min and discard the supernatant. Resuspend the cell pellet in 1 mL of cold MACS buffer by pipetting up and down.
Count the cells using a hemocytometer (or automated counting instrumentation if available). CRITICAL STEP It is important to calculate the cell number for determining the quantities of antibodies and MACS microbeads, as well as the size of MACS columns in the following steps. Normally, 5 × 107–7 × 107 total BMCs can be obtained from the hindlimbs of one mouse after RBC lysis.
Add another 4 mL of cold MACS buffer to the above cell suspension, centrifuge at 300 × g and 4°C for 5 min, and discard the supernatant. Resuspend the cell pellet in 100 μL of Dead Cell Removal MicroBeads per 107 cells. Mix well and incubate for 15 min at room temperature.
Apply the cell suspension in 1 mL of 1× DCB Buffer onto the pre-washed LS column in the magnetic field of the MACS separator. Collect the negative cells that passed through the column. Rinse the column with 3 mL of DCB Buffer thrice and collect all effluent as the live cell fraction (normally 70-80% of the cells from the last step could be collected after dead cell removal). Centrifuge the live cell suspensios at 300 × g and 4°C for 5 min, and discard the supernatant.
Resuspend the cell pellet in 1 mL of cold MACS buffer. Count the cells using a hemocytometer (or automated counting instrumentation if available), then proceed to primary antibody labeling.
Magnetic isolation of CTR+ and CTR– cells ● TIMING 2 h 20 min
Centrifuge the above cell suspension at 300 × g and 4°C for 5 min, and discard the supernatant. Then, resuspend the cell pellet with 2 μg of Biotin-conjugated CTR antibody and OSCAR antibody in 200 μL of MACS buffer per 107 cells. Incubate in the dark at 4°C for 30 min.
Wash the cell suspensions with 5 mL of cold MACS buffer and centrifuge at 300 × g and 4°C for 5 min. Repeat the washing steps twice. CRITICAL STEP To reduce the unbound primary antibody, it is important to wash the cells at least twice following antibody labeling.
Resuspend the cells with 20 μL of anti-Biotin MultiSort Microbeads in 80 μL of cold MACS buffer per 107 cells. Incubate in the dark at 4°C for 15 min.
Wash the cell suspension with 5 mL of cold MACS buffer and centrifuge at 300 × g and 4°C for 10 min. Repeat the washing steps twice and resuspend the cells in 500 μL of cold MACS buffer.
Proceed to magnetic separation. Insert the LS column with the column wings to the front into a MACS separator. Prepare the LS column by rinsing it with cold MACS buffer: apply 2 mL of degassed MACS buffer on top of the column and allow the MACS buffer to run through. Discard the effluent and change the collection tube. CAUTION! The columns are “flow stop” and do not run dry. The maximum sample size for an LS column is 1 × 108 labeled cells in 2 × 109 total cells.
Apply the cell suspensions onto the prepared LS column. Collect the effluent that passes through. Wash the LS column for thrice by adding 3 mL of degassed MACS buffer each time when the column reservoir is empty. Collect the total effluent containing the CTR– cells.
Remove the LS column from the separator and place it on a new collection tube. Pipette 5 mL of MACS buffer onto the LS column. Immediately flush out the fraction with magnetically labeled cells, by firmly applying the plunger supplied with the column. CAUTION! The flushed cells in this step are the CTR+ cells. To increase the purity of magnetically separated CTR+ cells, repeat the magnetic separation procedure as described in Steps 5–7 with a new column.
Centrifuge the CTR– cell fraction in Step 6 and the CTR+ cell fraction in Step 7 at 300 × g and 4°C for 10 min. Discard the supernatants, and proceed to the next step for eluting the magnetic beads.
Magnetic separation of OSCAR+CTR+ and OSCAR+CTR– cells ● TIMING 2 h
Resuspend the above cell pellet of each cell fraction with 1 μL of MultiSort Release Reagent in 100 μL of cold MACS buffer per 107 cells. Mix well and incubate at 4°C for 10 min. Wash the cells with 1 mL of the cold MACS buffer per 107 cells and centrifuge at 300 × g and 4°C for 10 min. Discard the supernatant and resuspend the cells in 50 μL of cold MACS buffer per 107 cells. Add 30 μL of MultiSort Stop Reagent per 107 cells into the cell suspensions and mix well. Centrifuge the above cell suspensions at 300 × g and 4°C for 5 min. CRITICAL STEP The anti-Biotin MultiSort microbeads are removed from the cells at this step, which allows for a second-round of magnetic separation of the OSCAR+ cells.
Resuspend the above cell pellet of each cell fraction in 1 mL of cold MACS buffer. Count the cells using a hemocytometer (or automated counting instrumentation if available). Centrifuge the above cell suspensions at 300 × g and 4°C for 5 min and discard the supernatants. Then, resuspend the cell pellets with 20 μL of anti-Rat IgG Microbeads in 80 μL of cold MACS buffer per 107 cells and incubate in the dark at 4°C for 15 min.
Wash the cell suspensions with 5 mL of cold MACS buffer and centrifuge at 300 × g and 4°C for 10 min. Repeat the washing steps twice, and resuspend the cells in 500 μL of cold MACS buffer.
Proceed to the magnetic separation. Insert the MS column with the column wings to the front into a MACS separator. Prepare the MS column by rinsing with cold MACS buffer: apply 500 μL of degassed MACS buffer on top of the column and let the MACS buffer run through. Discard the effluent and change the collection tube. CAUTION! The columns are “flow stop” and do not run dry. The maximum sample size for an MS column is 1 × 107 labeled cells in 2 × 108 total cells.
Apply the CTR+ and CTR– fractions cell suspensions onto the prepared MS column. Collect the effluent that passes through. Wash the MS column thrice by adding 500 μL of degassed MACS buffer each time when the column reservoir is empty. Collect the total effluent, which contains the OSCAR– cells (non-OLCs).
Remove the MS column from the separator and place it on a new collection tube. Pipette 1 mL of MACS buffer onto the MS column. Immediately flush out the fraction with magnetically labeled cells, by firmly applying the plunger supplied with the column. CAUTION! The flushed cells in this step are the OSCAR+CTR+ (MOCs) and OSCAR+CTR– cells (OCPs), respectively.
Centrifuge the desired cell fractions at 300 × g and 4°C for 10 min to pellet the cells, and discard the supernatant. The cell pellets can proceed to RNA extraction for gene expression assays, e.g., RT-PCR and microarray, or protein extraction for western blot. Alternatively, resuspend the cell pellets in appropriate cell culture medium for osteoclastogenic cell culture, followed by tartrate-resistant acid phosphatase (TRAP) staining and bone resorption assay, for measuring the osteoclast activity in vitro. CRITICAL STEP Trypan blue staining is performed for counting viable cells.
Data analysis
Enrichment of the MACS-sorted OCPs and MOCs
The purity of OSCAR+ CTR– and OSCAR+ CTR+ cells in the MACS-sorted cell fractions were generally >75% and >90%, respectively (Figure 2). The enrichment of these two cell fractions after MACS was high and reproducible in both OVX and Sham mice (Supplementary Figure 1a).
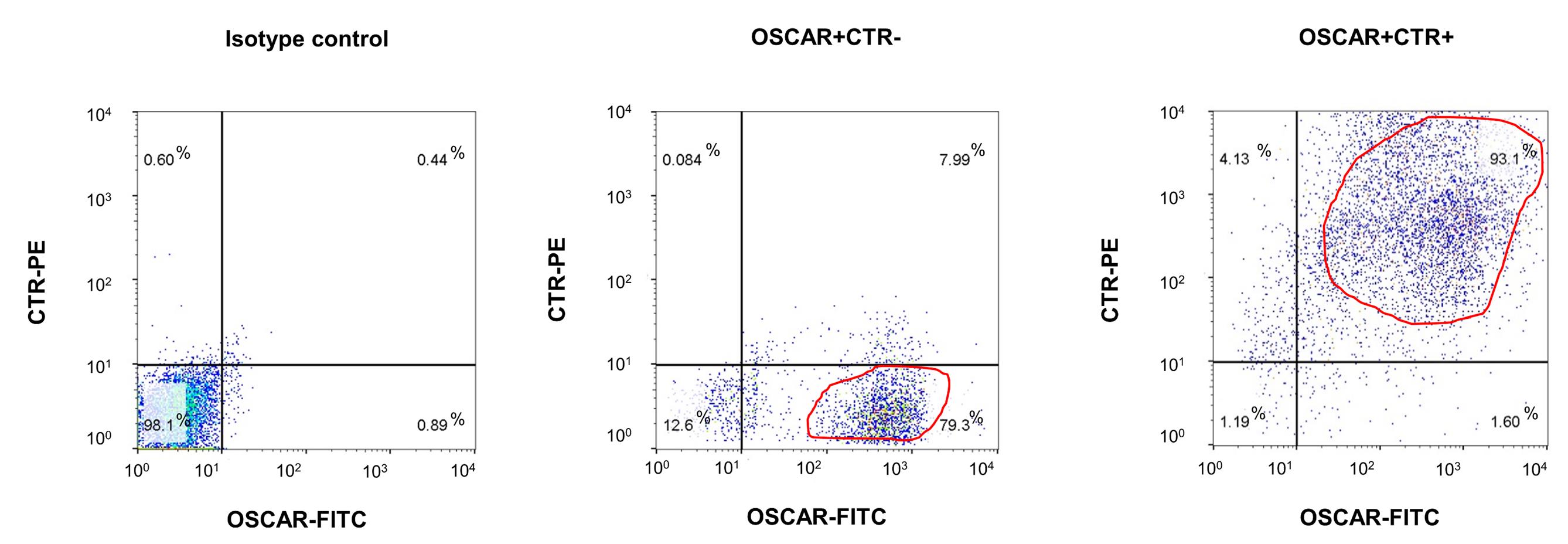
Figure 2.The purity of MACS-sorted OSCAR+CTR– and OSCAR+CTR+ cells. Flow cytometry analysis showing the purity of MACS-sorted OSCAR+CTR– and OSCAR+CTR+ cells. The dead cells were excluded by 7-aminoactinomycin D (7-AAD) staining, while the live cells were gated for detecting the expression of OSCAR (labelled by FITC-conjugated secondary antibody) and CTR (labelled by PE-conjugated secondary antibody) in MACS-sorted OSCAR+CTR– osteoclast precursors (middle panel) and MACS-sorted OSCAR+CTR+ mature osteoclasts (right panel). Isotype control antibodies were used as negative controls (left panel).Molecular and functional characterization of the MACS-sorted OCPs and MOCs
The expression of tartrate-resistant acid phosphatase (TRAP, encoded by the ACP5 gene) defines the molecular features of OLCs. Together with high expression of Cathepsin K (CTSK), these are important markers of osteoclasts maturation. Consistently, the real-time qPCR and western blot analysis (details in Supplementary Methods) showed that the mRNA and protein expression levels of the osteoclastic markers TRAP and CTSK in the OSCAR+ CTR– and OSCAR+ CTR+ cells were both remarkably higher than those in the OSCAR– cell fractions (Figure 3a–b). Moreover, the mRNA and protein expression levels of TRAP and CTSK were both significantly higher in the OSCAR+ CTR+ cells when compared to the OSCAR+ CTR– cells (Figure 3a–b). On the other hand, the mature osteoclasts are characterized as TRAP-stained multinucleate cells with activated bone resorbing capacity, while osteoclast precursors are characterized as TRAP-stained mononucleate cells without bone resorbing capacity. Consistently, the TRAP staining and bone resorption assay (details in Supplementary Methods) revealed that the OSCAR– cells were homogeneously TRAP-negative without bone resorbing capacity, the OSCAR+CTR– cells are mostly TRAP-positive mononucleate cells with little bone resorbing capacity, while the OSCAR+CTR+ cells are TRAP-positive multinucleate cells with high bone resorbing capacity (Figure 4a–b).
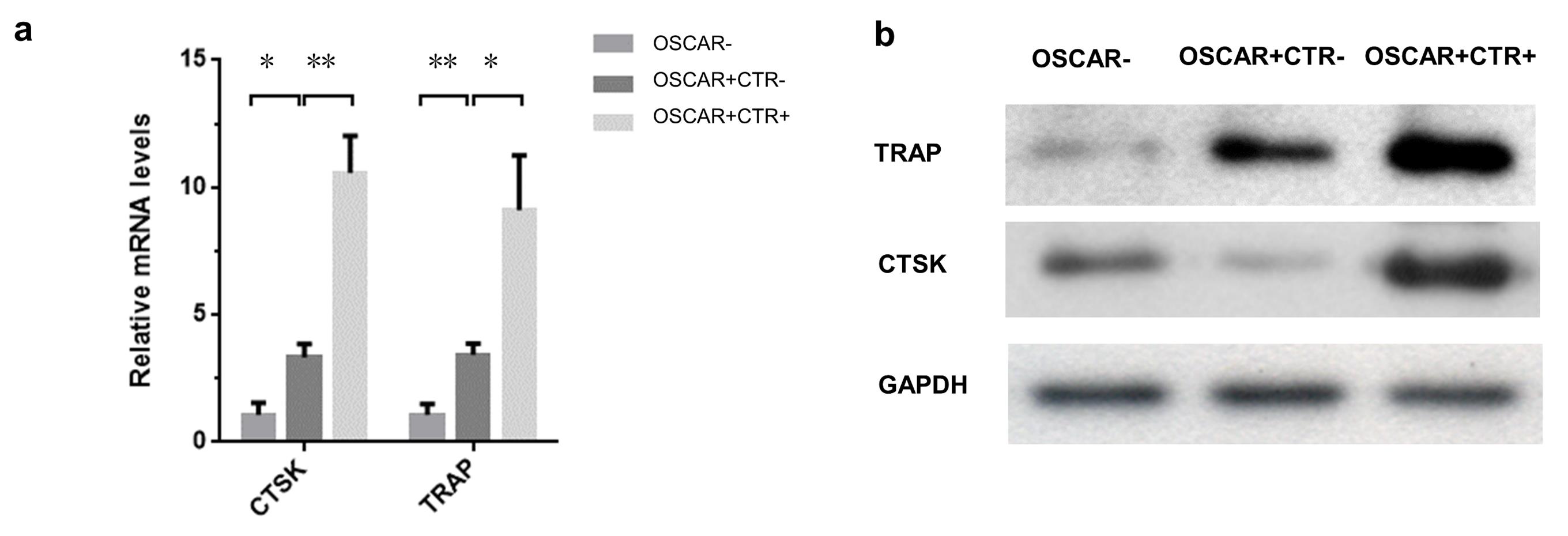
Figure 3. The molecular characterization of the MACS-sorted OSCAR–, OSCAR+CTR–, and OSCAR+CTR+ cells from the Sham mice. (a) Real-time qPCR analysis of TRAP and CTSK mRNA expression in the OSCAR–, OSCAR+CTR–, and OSCAR+CTR+ cells. (b) Western blot analysis of the TRAP and CTSK expression in the OSCAR–, OSCAR+CTR–, and OSCAR+CTR+ cells. All data are the mean ± S.D. *P < 0.05, **P < 0.01. A one-way ANOVA with subsequent Tukey's multiple comparisons test was performed.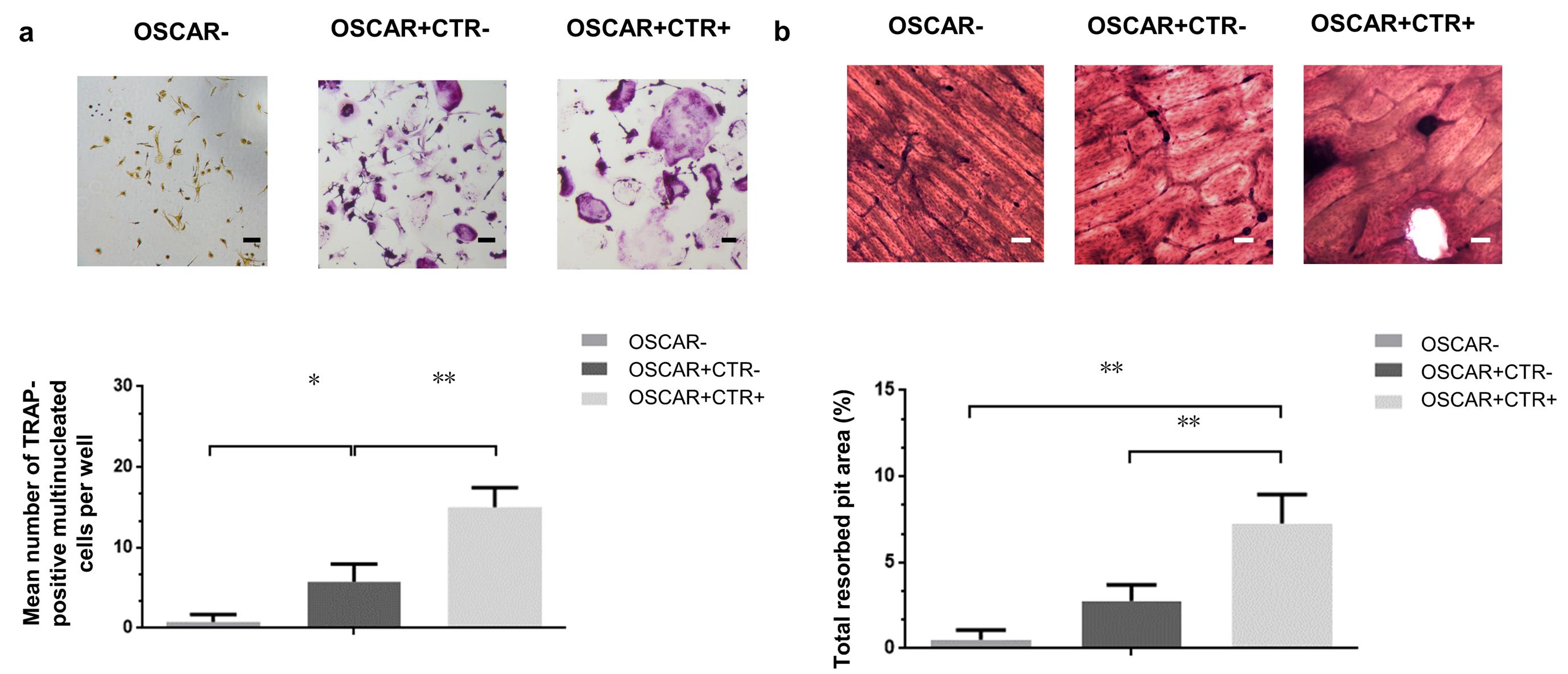
Figure 4. The functional characterization of the MACS-sorted OSCAR–, OSCAR+CTR–, and OSCAR+CTR+ cells from the Sham mice. (a) TRAP staining of the OSCAR–, OSCAR+CTR–, and OSCAR+CTR+ cells. The TRAP-positive cells with more than three nucleic were counted. Scale bars, 100 μm. (b) Bone-resorptive assay of the OSCAR–, OSCAR+CTR–, and OSCAR+CTR+ cells. Scale bars, 100 μm. All data are the mean ± S.D. *P < 0.05, **P < 0.01. A one-way ANOVA with subsequent Tukey's multiple comparisons test was performed.Pathological changes of the MACS-sorted OCPs and MOCs after ovariectomy
The ovariectomy-induced estrogen depletion could result in aberrantly increased osteoclast formation with excessive bone resorption, leading to significant bone loss in mice. Consistently, the mRNA and protein expression levels of TRAP and CTSK were dramatically higher in the OSCAR+CTR– and OSCAR+CTR+ cells isolated from OVX mice compared to those cells isolated from Sham mice (Supplementary Figure 1b-c). Moreover, the TRAP-positive mononucleate OSCAR+CTR– cells isolated from OVX mice developed more TRAP-positive multinucleate cells after stimulation with the Receptor activator of nuclear factor kappa-&Bgr; ligand (RANKL) compared to those isolated from Sham mice. Similarly, the TRAP-positive multinucleate OSCAR+CTR+ cells isolated from the OVX mice presented superior bone resorbing capacity than those isolated from Sham mice (Supplementary Figure 1d-e).
In conclusion, this article provides an easy-to-operate protocol for rapidly isolating the live OCPs and MOCs directly from mouse BMCs in vivo. The enriched live cells obtained through this protocol could truly retain their in vivo status, which would be valuable for studying the biological features and pathological changes of OLCs in vivo. This protocol also shows promise if employed for isolating the live OLCs from mice of various species, age, and gender.
Notes
Problem | Possible reason | Solution |
Low purity (OSCAR+CTR– cells < 60%, OSCAR+CTR+ cells < 80%) | Numbers of the magnetically labelled cells exceeded MS column binding capacity | Reload the positive fraction into the new column |
Unspecific binding in the column | Use MACS buffer to wash column more times and reload the positive fraction into the new column | |
Low yield (OSCAR+CTR– cell Number < 5 × 106 per mouse, OSCAR+CTR+ cell Number < 1 × 105 per mouse) | Too many cells remaining in the bone marrow | Flush out of bone marrow cells as much as possible |
Cells are incubated in the red blood cell lysis for too long time, leading to a lot of cell debris. | Strict control of the lysis time | |
Washing of unbound antibody was insufficient | Wash cell suspensions twice with 10 volumes of MACS buffer | |
Many dead cells | Keep cell suspensions on ice and carry out all procedures with cold reagents |
Recipes
Complete DMEM Medium
Mix DMEM with 10% (vol/vol) of FBS and 1% (vol/vol) of penicillin/streptomycin. Prepare in advance and store at 4°C for up to 1 month.
DPBS (1×)
1× DPBS is prepared using 80 g NaCl, 2.0 g KCl, 11.5 g Na2HPO4, 2.0 g KH2PO4, and 800 mL of distilled H2O. Adjust pH to 7.4 and add distilled H2O to adjust the volume to 1 L. Prepare in advance, autoclave, and store at room temperature (21–24 °C) for up to 6 months.
1× RBC Lysis Solution
Mix 10 mL of 10× Red Blood Cell Lysis Solution and 90 mL of distilled H2O. It is not necessary to prepare the solution in advance.
1× DCB buffer
Dilute the 20× DCB buffer stocking solution with sterile double distilled H2O, to prepare 1× DCB buffer.
MACS buffer
Mix 0.37 g EDTA and 2.5 g BSA, add 1× DPBS to a volume of 500 mL, and filter-sterilize and degas the solution. Prepare in advance and store at 4°C for up to 1 month.
1% Toluidine Blue solution
Mix 1 g Toluidine blue into 100 mL of 70% ethanol. Add 1× DPBS to adjust the volume to 1,000 mL and mix well. The pH should be around 2.3 and less than 2.5. Prepare in advance and store at 4°C for up to 1 month.
Acknowledgments
This work was supported by the funds from National Key R&D Program of China (2018YFA0800804 to L.C.) and National Natural Science Foundation Council of China (81702189 to J. L), the Theme-based Research Scheme from the Research Grants Council of Hong Kong (T12-201/20-R to A.L.), the General Research Funds from the Research Grants Council of Hong Kong (12100918, 12114416 and 12101117 to G.Z., 12136616 and 12103519 to J. L, 14112915 to B.-T.Z.), the Basic and Applied Basic Research Fund from Department of Science and Technology of Guangdong Province (2019B1515120089 to G.Z.), the 2020 Guangdong Provincial Science and Technology Innovation Strategy Special Fund (Guangdong-Hong Kong-Macau Joint Lab, 2020B1212030006),the Interdisciplinary Research Clusters Matching Scheme of Hong Kong Baptist University (RC-IRCs/17-18/02 to G.Z.; RC-IRCs/17-18/04 and RC-IRMS/15-16/01 to A.L.), the Inter-institutional Collaborative Research Scheme from Hong Kong Baptist University (RC-ICRS/19-20/01 to A.L.). This protocol was modified from previous work (Liu et al., 2015 and 2021).
Competing interests
The authors declare no conflict of interest.
Ethics
All the experimental procedures were approved by the Committees of Animal Ethics and Experimental Safety of Hong Kong Baptist University and the Chinese University of Hong Kong.
References
- Almeida, M., Garcia-Montero, A. C. and Orfao, A. (2014). Cell Purification: A New Challenge for Biobanks. Pathobiology 81(5-6): 261-275.
- Asagiri, M. and Takayanagi, H. (2007). The molecular understanding of osteoclast differentiation. Bone 40(2): 251-264.
- Atkins, G. J., Kostakis, P., Vincent, C., Farrugia, A. N., Houchins, J. P., Findlay, D. M., Evdokiou, A. and Zannettino, A. C. W. (2006). RANK expression as a cell surface marker of human osteoclast precursors in peripheral blood, bone marrow, and giant cell tumors of bone. J Bone Miner Res 21(9): 1339-1349.
- Collin-Osdoby, P. and Osdoby, P. (2012). Isolation and culture of primary chicken osteoclasts. Methods Mol Biol 816: 119-143.
- Elefteriou, F. and Yang, X. L. (2011). Genetic mouse models for bone studies-Strengths and limitations. Bone 49(6): 1242-1254.
- Feng, X. and McDonald, J. M. (2011). Disorders of Bone Remodeling. Abbas, A. K., Galli, S. J. and Howley, P. M. (Eds.) Annual Review of Pathology: Mechanisms of Disease, Vol 6. Annual Reviews, 121-145.
- Glantschnig, H., Fisher, J. E., Wesolowski, G., Rodan, G. A. and Reszka, A. A. (2003). M-CSF, TNFalpha and RANK ligand promote osteoclast survival by signaling through mTOR/S6 kinase. Cell Death Differ 10(10): 1165-1177.
- Lee, M. Y. and Lufkin, T. (2012). Development of the"Three-step MACS": a novel strategy for isolating rare cell populations in the absence of known cell surface markers from complex animal tissue. J Biomol Tech 23(2): 69-77.
- Li, D., Liu, J., Guo, B., Liang, C., Dang, L., Lu, C., He, X., Cheung, H. Y., Xu, L. and Lu, C. (2016). Osteoclast-derived exosomal miR-214-3p inhibits osteoblastic bone formation. Nat Commun 7: 10872.
- Liu, J., Dang, L., Li, D., Liang, C., He, X., Wu, H., Qian, A., Yang, Z., Au, D. W. and Chiang, M. W. (2015). A delivery system specifically approaching bone resorption surfaces to facilitate therapeutic modulation of microRNAs in osteoclasts. Biomaterials 52: 148-160.
- Liu, J., Wu, X., Lu, J., Huang, G., Dang, L., Zhang, H., Zhong, C., Zhang, Z., Li, D. and Li, F. (2021). Exosomal transfer of osteoclast-derived miRNAs to chondrocytes contributes to osteoarthritis progression. Nature Aging 1(4): 368-384.
- Madel, M. B., Ibanez, L., Rouleau, M., Wakkach, A. and Blin-Wakkach, C. (2018). A novel reliable and efficient procedure for purification of mature osteoclasts allowing functional assays in mouse cells. Front Immunol 912.
- Marino, S., Logan, J. G., Mellis, D. and Capulli, M. (2014). Generation and culture of osteoclasts. Bonekey Rep 3: 570.
- Novack, D. V. and Mbalaviele, G. (2016). Osteoclasts-Key players in skeletal health and disease. Microbiol Spectr 4(3): 19.
- Quinn, J. M. W., Neale, S., Fujikawa, Y., McGee, J. O. D. and Athanasou, N. A. (1998). Human osteoclast formation from blood monocytes, peritoneal macrophages, and bone marrow cells. Calcif Tissue Int 62(6): 527-531.
- Zhang, G., Guo, B. S., Wu, H., Tang, T., Zhang, B. T., Zheng, L. Z., He, Y. X., Yang, Z. J., Pan, X. H. and Chow, H. (2012). A delivery system targeting bone formation surfaces to facilitate RNAi-based anabolic therapy. Nat Med 18(2): 307-314.
- Zhen, G. H., Wen, C. Y., Jia, X. F., Li, Y., Crane, J. L., Mears, S. C., Askin, F. B., Frassica, F. J., Chang, W. Z. and Yao, J. (2013). Inhibition of TGF-beta signaling in mesenchymal stem cells of subchondral bone attenuates osteoarthritis. Nat Med 19(6): 704-712.
- 18. Zhu, S. A., Zhu, J. X., Zhen, G. H., Hu, Y. H., An, S. B., Li, Y. S., Zheng, Q., Chen, Z. Y., Yang, Y. and Wan, M. (2019). Subchondral bone osteoclasts induce sensory innervation and osteoarthritis pain. J Clin Invest 129(3): 1076-1093.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Dang, L., Li, N., Wu, X., Li, D., Zhang, Z., Zhang, B. T., Lyu, A., Chen, L., Zhang, G. and Liu, J. (2022). A Rapid Protocol for Direct Isolation of Osteoclast Lineage Cells from Mouse Bone Marrow. Bio-protocol 12(5): e4338. DOI: 10.21769/BioProtoc.4338.
Category
Cell Biology > Cell isolation and culture > Cell isolation > Dynabead
Biological Engineering > Biomedical engineering
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









