- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Amber Suppression Technology for Mapping Site-specific Viral-host Protein Interactions in Mammalian Cells
Published: Vol 12, Iss 3, Feb 5, 2022 DOI: 10.21769/BioProtoc.4315 Views: 4209
Reviewed by: Khyati Hitesh ShahHeng SunWenn-Chyau Lee

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Analysis of in vivo Interaction between RNA Binding Proteins and Their RNA Targets by UV Cross-linking and Immunoprecipitation (CLIP) Method
Pamela Bielli and Claudio Sette
May 20, 2017 11523 Views

Cross-linking, Immunoprecipitation and Proteomic Analysis to Identify Interacting Proteins in Cultured Cells
Hao Wang [...] Qingyu Wu
Jun 5, 2019 15241 Views
DSP-crosslinking and Immunoprecipitation to Isolate Weak Protein Complex
Kotaro Akaki [...] Osamu Takeuchi
Aug 5, 2022 8702 Views
Abstract
Probing the molecular interactions of viral-host protein complexes to understand pathogenicity is essential in modern virology to help the development of antiviral therapies. Common binding assays, such as co-immunoprecipitation or pull-downs, are helpful in investigating intricate viral-host proteins interactions. However, such assays may miss low-affinity and favour non-specific interactions. We have recently incorporated photoreactive amino acids at defined residues of a viral protein in vivo, by introducing amber stop codons (TAG) and using a suppressor tRNA. This is followed by UV-crosslinking, to identify interacting host proteins in live mammalian cells. The affinity-purified photo-crosslinked viral-host protein complexes are further characterized by mass spectrometry following extremely stringent washes. This combinatorial site-specific incorporation of a photoreactive amino acid and affinity purification-mass spectrometry strategy allows the definition of viral-host protein contacts at single residue resolution and greatly reduces non-specific interactors, to facilitate characterization of viral-host protein interactions.
Graphic abstract:
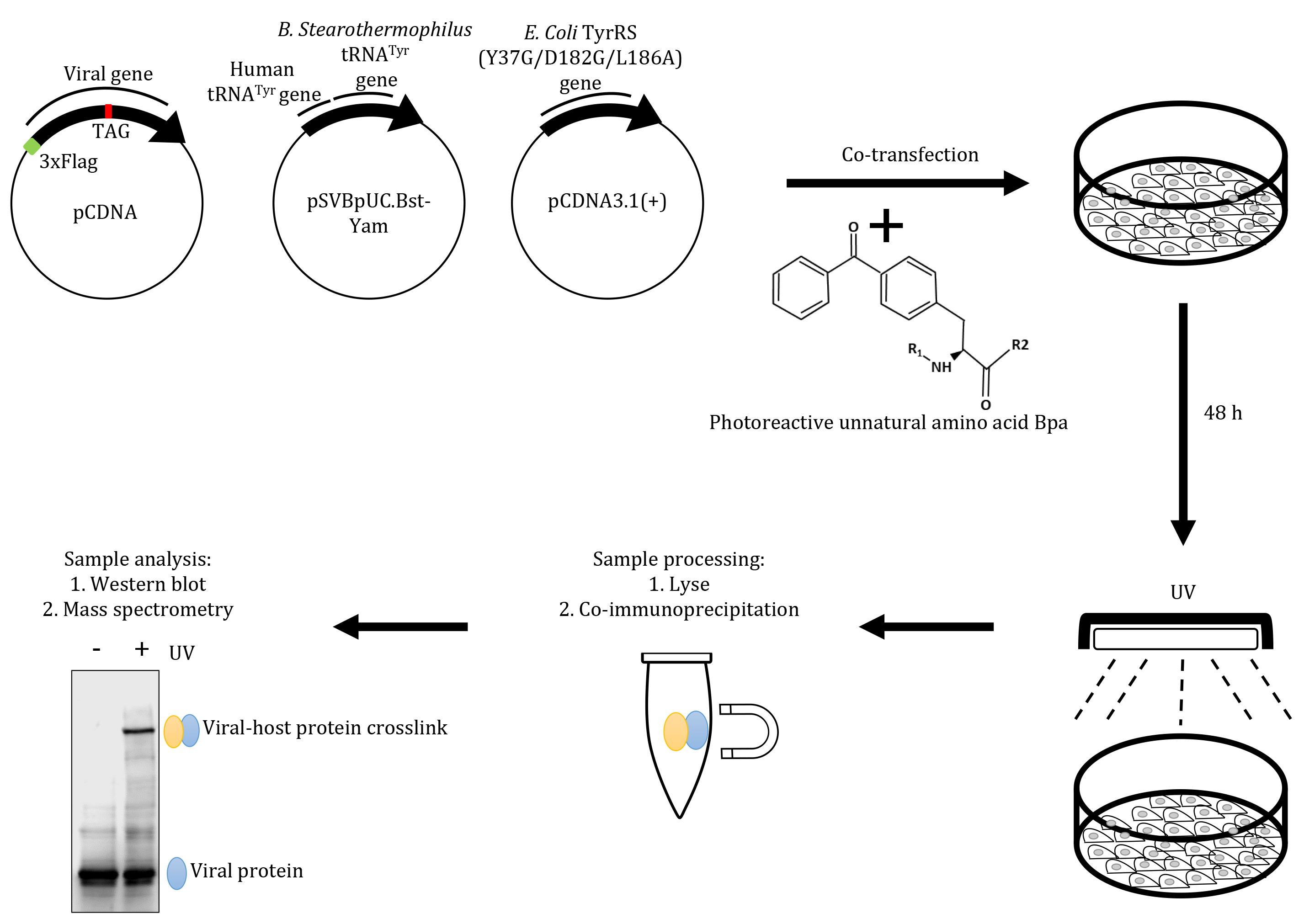
Schematic overview of the virus-host interaction assay based on an amber suppression approach. Mammalian cells grown in Bpa-supplemented medium are co-transfected with plasmids encoding viral sequences carrying a Flag tag, a (TAG) stop codon at the desired position, and an amber suppressor tRNA (tRNACUA)/aminoacyl tRNA synthetase (aaRS) orthogonal pair. Cells are then exposed to UV, to generate protein-protein crosslinks, followed by immunoprecipitation with anti-Flag magnetic beads. The affinity-purified crosslinks are probed by western blot using an anti-Flag antibody and the crosslinked host proteins are characterised by mass spectrometry.
Background
In situ binding assays such as pull-downs or affinity purification coupled with mass spectrometry are reliable approaches to study viral-host protein interactions. However, it is important to recognize the limitations of these methods, including lack of specificity, which can lead to false-positives in the interactome. To minimize non-specific binding artifacts, we have designed an interaction assay based on ectopic expression of an engineered viral protein in mammalian cells, in the presence of a suppressor tRNA (tRNASup)/aminoacyl tRNA synthetase (aaRS) orthogonal pair and growth medium supplemented with an unnatural photoreactive amino acid (Ye et al., 2008; Neumann-Staubitz and Neumann, 2016; Xiao and Schultz, 2016; Chin, 2017). The amber codon TAG is first introduced at the desired residues within the viral protein. During ectopic expression of these mutated viral proteins in the presence of the tRNASup/aaRS pair in mammalian culture, the aaRS charges an unnatural photocrosslinkable amino acid to the tRNASup, which is then incorporated at the position of the amber codon. The cells expressing the viral protein containing the unnatural amino acid are then exposed to UV light, which induces covalent photocrosslinks to host proteins present in close proximity to the viral protein. The crosslinked complex is stable and can be affinity-purified in stringent conditions, to eliminate non-specific interactions and be characterised by mass spectrometry. Over the years, the approach has been applied to interactions between cellular proteins (Hino et al., 2005; Kobbi et al., 2016; Wu et al., 2020). We have adapted this approach to interrogate the interaction between host-cell proteins with the viral protein HSV-1 ICP22 (Isa et al., 2021), and this is applicable to other viral proteins.
Materials and Reagents
1.5 mL microcentrifuge tube (Starlab, catalog number: E1415-1510)
Pipette tip (10 μL, 200 μL, and 1,000 μL) (Sarstedt, catalog numbers: 70.3010, 70.760.3010, 70.3050.200)
Cell culture Petri dishes (Starlab, catalog number: CC7682-3394)
Mammalian expression vector carrying N-terminally 3× FLAG-tagged proteins, as for example p3× FLAG-CMVTM-8 (Merck, catalog number: E9908)
pSVBpUC.Bst-Yam suppressor tRNA plasmid, a generous gift from Dr. Shixin Ye-Lehman (read as tRNASup). Gene cloning was described by Ye et al. (2008)
pCDNA3.1(+).aminoacyl-tRNA synthetase specific for Bpa, a generous gift from Dr. Shixin Ye-Lehman (read as aaRS). Gene cloning was described by Ye et al. (2008)
HEK293 cells (ATCC®, catalog number: CRL-1573TM)
Anti-Flag M2 Magnetic beads (Sigma-Aldrich, catalog number: M8823; -20°C)
Product H-L-Bpa-OH (p-benzoyl-L-phenylalanine) (IRIS Biotech, catalog number: HAA6010) (read as Bpa)
QuikChange multi site-directed mutagenesis kit (Agilent Technologies, catalog number: 200514)
DMEM (Sigma-Aldrich, catalog number: D6429; 4°C)
jetPRIME® transfection reagent (PolyPlus, catalog number: 114-15; 4°C)
cOmpleteTM EDTA-free Protease Inhibitor Cocktail (Sigma-Aldrich, catalog number: 5056489001, 4°C). Store dissolved stock solution at -20°C
Benzonase (Merck Chemicals, catalog number: 70664-3; -20°C)
Anti-flag M2 antibody (Sigma Aldrich, catalog number: F1804; -20°C)
PBS (Lonza, catalog number: BE17-516F)
NaOH (VWR Chemicals, catalog number: 28244.262)
HCl (Fisher ChemicalTM, catalog number: H/1150/PB17)
Foetal bovine serum (Gibco, catalog number: 10500064; -20°C)
Penicillin-streptomycin (Gibco, catalog number: 15140-122; 4°C)
L-glutamine (Gibco, catalog number: 25030024, 4°C)
HEPES (Sigma-Aldrich, catalog number: H0887)
KCl (Sigma-Aldrich, catalog number: P9541)
NaCl (VWR Chemicals, catalog number: 27810.364)
EDTA (VWR Chemicals, catalog number: 20302.260)
EGTA (Sigma-Aldrich, catalog number: E3889)
NP-40 (IGEPAL® CA-630)( Sigma-Aldrich, catalog number: I3021)
Urea (VWR Chemicals, catalog number: 28877.292)
Sodium deoxycholate (Sigma-Aldrich, catalog number: D6750)
SDS (Sigma-Aldrich, catalog number: 05030)
Precision Plus protein dual color standard (Bio-Rad, catalog number: 1610374)
Gentle RIPA buffer (see Recipes)
Stringent RIPA buffer (see Recipes)
Equipment
Benchtop chilled centrifuge (Thermo Scientific, model: HeraeusTM Fresco 17 or MicroCL 21R)
UV lamp (UVP, model: 95-0007-06)
DNA Shearing Sonicator (Diagenode, model: Bioruptor® Pico or QSonica, model: Q800R1)
Pipette controller (Starlab, model: Starpet Pro E4866-0021)
Tube rotator (Stuart, model: SB3)
Standard orbital shaker (Stuart, model: SSL3)
Procedure
Design and construction of plasmid vectors for the wild type and TAG-mutated viral protein
Clone a gene encoding the viral protein into a vector encoding an N-terminal 3× FLAG tag.
Perform cell transformation, plasmid purification, and confirmation by DNA sequencing according to standardized protocols in individual labs.
Design primers to replace a selected codon of the wild type viral gene with the amber codon TAG, ideally located in the centre of the both forward and reverse primers.
Note: First select aromatic amino acids such as tyrosine, phenylalanine, and tryptophan (The Bpa resembles tryptophan). Other residues such as aliphatic amino acids (isoleucine, or leucine) may also be replaced. Prioritize those within evolutionary-conserved regions.
Perform oligonucleotide-based site-directed mutagenesis using QuikChange multisite-directed mutagenesis kit (Agilent Technologies) according to the manufacturer’s instructions.
Preparation of the Bpa solution
Freshly prepare a Bpa solution by first dissolving Bpa (Molecular weight: 269.29 g/mol) in 1 M NaOH in a weighing boat. Example: 54 mg of Bpa (Molecular weight: 269.29 g/mol) in 220 µL of NaOH solution for 100 mL of culture medium. This mother solution should be limpid.
Note: Prepare an adequate amount of Bpa solution every time prior to experiments. Preparation can be at room temperature without protection from light. Sterilization is not necessary for the NaOH solution or Bpa in the NaOH solution. Leave the weighing boat covered with cling film on an orbital shaker. Bpa takes a few minutes to completely dissolve resulting in a yellow-greenish solution.
Then, add the Bpa solution to 100 mL of culture medium to obtain a final concentration of 2 mM.
Gently mix the medium.
Add 220 µL of 1 M HCl to neutralize the culture medium (pH 6.8-7.2). Gently mix the medium again.
Co-expression
Note: For each new mutant derivative, it is advisable to first perform co-expression in 6-well plates and validate the incorporation of Bpa by western blot.
Add the Bpa-containing medium to HEK293 cells at 70-80% confluency.
Note: Prepare sets of Petri dishes with wild type or mutated derivatives in the absence or presence of Bpa.
Quantify all plasmid vectors before transfection. Use standard transfection reagents, such as jetPRIME® transfection reagent (PolyPlus) to co-transfect the three plasmid vectors encoding the viral gene, tRNASup, and aaRS, at a 1:2:1 ratio.
Note: The optimal ratio may change depending on the transfection efficiency and how well the proteins are expressed. However, these ratios worked in HEK293 cells for ICP22.
Add transfection solution to the side of the plate, gently rock the dishes, and place the cells back in the incubator.
After 24 h, replace medium with fresh Bpa-containing medium. Grow overnight.
Lyse cells and affinity purify the viral protein for western blot to screen the mutant derivatives for expression (Figure 1).
Note: Follow Section E for affinity purification.
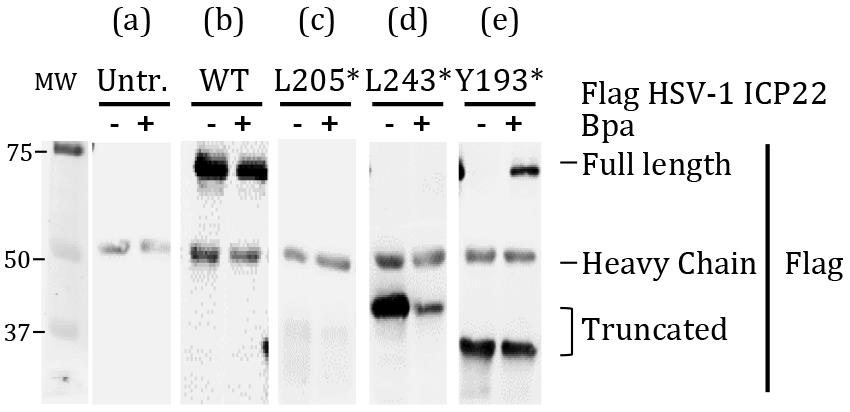
Figure 1. Affinity-purified HSV-1 ICP22 viral protein from HEK293 cells cultured with and without Bpa. Wild type (WT) has no amber codons in the gene. (a) In untransfected (Untr.) cells, the bands seen at about 55 kDa is the IgG heavy chain. Specific western blot signals are obtained from immunoprecipitated samples. (b) Protein extracts from cells expressing full-length wild-type viral protein without any amber codons migrates at about 70 kDa in the presence or absence of Bpa. The wild type viral protein serves as a point of reference for selection of good mutant derivatives. (c) Protein extracts from cells co-transfected with mutated viral protein L205TAG(*). Blot looks like (a), which indicates a failure of expression. Exclude these mutants from downstream experiments. (d) Protein extracts from cells co-transfected with mutated viral protein L243TAG(*). The expressed truncated viral protein migrates at about 40 kDa. However, the mutant failed to incorporate Bpa. Exclude from downstream experiments. (e) Protein extracts from cells co-transfected with mutated viral protein Y193TAG(*). The expressed truncated protein migrates at about 30 kDa. A portion of the truncated viral protein incorporates Bpa and full-length viral protein migrating at about 70 kDa is detected [corresponding to (b)]. Select derivatives that display this pattern for live cell UV crosslinking.
Live cell UV crosslinking
Note: Prepare sets of Petri dishes including non UV-irradiated versus UV-irradiated wild-type or mutated derivatives of the viral gene. Experiments are performed in 10-cm dishes to obtain sufficient crosslinked products for immunoprecipitation and mass spectrometry.
Gently remove the culture medium.
Briefly wash off any residue with 5 mL of room-temperature PBS.
Gently add 5 mL of PBS to the side of the plate.
For UV crosslinking, the experiment is ideally performed in a cold room, as sterility is not necessary from this step onwards, although clean conditions are.
Place the cells horizontally and stably on top of a chilled metal plate.
Note: Alternatively, place the cells on ice, but ice needs to be frequently changed due to prolonged UV exposure causing it to melt.
Place a clean handheld UV irradiator on top of lid-less Petri dishes. Set the irradiator to 365 nm UV and completely cover the irradiator and the Petri dishes with aluminum foil. Irradiate the cells for 45 min.
Note: A portable handheld UV irradiator is convenient. However, it can only accommodate a limited number of 10-cm dishes at a time. A benchtop UV irradiator can be used, but it may have a different UV intensity. In this case, check exposure time every 20 min to see if cells remain viable.
Lysis and fractionation
Note: Two lysis buffers are used: the stringent RIPA buffer for total cell lysis or nuclear lysis, and the gentle RIPA buffer for cytoplasmic fractionation. Pre-equilibrate anti-Flag M2 beads by washing with the respective buffers 2-3 times, at 5 min intervals.
Work on ice.
Detach UV-irradiated cells and control non-UV irradiated cells by gentle scraping.
Note: If cells are detached, collect by gentle pipetting.
Spin the suspension at 200 × g at 4°C for 3 min.
Remove PBS completely.
Resuspend the cell pellets in 900 μL of gentle lysis buffer, to obtain the cytoplasmic fraction. Alternatively, resuspend the cell pellets in 900 μL of stringent lysis buffer, to obtain total cell lysates and proceed to step 10.
Add 1× protease inhibitors and homogenize by pipetting.
Note: Avoid frothing.
Spin down at 12,000 × g at 4°C for 10-15 min.
Transfer the non-pelleted solution (the cytoplasmic fraction) into 15 μL of pre-equilibrated beads in a fresh microcentrifuge tube for immunoprecipitation. Alternatively, freeze the recovered cytoplasmic fraction at -20°C until later.
Resuspend the nuclear pellet in 900 μL of stringent lysis buffer.
Sonicate for 10 cycles of 10 s on/off in a chilled water bath.
Note: Avoid frothing.
Add 8 μL of benzonase, rotate for 30 min in cold room for 1 h or until cell lysate viscosity is reduced.
Spin down at 12,000 × g at 4°C for 10-15 min.
Add the nuclear fraction or the total lysates to pre-equilibrated beads in a fresh microcentrifuge tube for immunoprecipitation. Alternatively, freeze the recovered supernatant at -20°C until further use.
Incubate the beads overnight at 4°C on a tube rotator.
Crosslink analysis
Wash the beads with the respective buffer.
Perform SDS-PAGE and western blot according to standardized protocols in individual labs. Probe with anti-Flag M2.
Note: For proteins that may migrate the same as heavy and light chain, perform Flag elution prior to western blot. Smaller proteins can be resolved by SDS-PAGE with Tris Tricine-SDS running buffer.
Analyse results as follows (Figure 2):
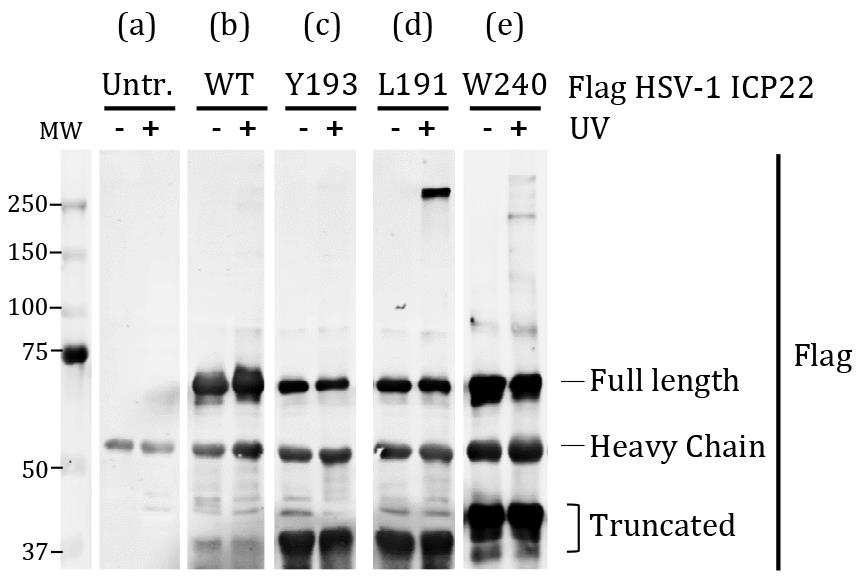
Figure 2. Affinity-purified HSV-1 ICP22 viral protein from HEK293 cells with or without UV exposure. (a) In untransfected (Untr.) HEK293 cells, the band seen at about 50 kDa is the IgG heavy chain. The western blot signals from immunoprecipitated samples are specific. (b) Protein extracts from cells expressing full-length wild-type (WT) viral protein, which migrates at about 70 kDa with or without UV irradiation. The wild-type viral protein serves as a point of reference for identification of crosslinked protein complexes. (c) Protein extracts from cells co-transfected with mutated viral protein Y193Bpa. Blot is comparable to (b) which indicates no visible crosslinked protein complex. Exclude these mutants from downstream analysis. (d) Protein extracts from cells co-transfected with mutated viral protein L191Bpa. A crosslink product is visible at about 250 kDa. Select derivatives that display this pattern for mass spectrometry. (e) Protein extracts from cells co-transfected with mutated viral protein W240Bpa. Several crosslinked products are visible at about 150-250 kDa. Derivatives that display this pattern are promising and can be considered for mass spectrometry.
Recipes
Gentle RIPA buffer
10 mM HEPES
10 mM KCl
200 mM NaCl
0.1 mM EDTA
0.1 mM EGTA
0.5% NP-40
Protease inhibitor cocktail
Stringent RIPA buffer
50 mM HEPES
1 M NaCl
1 mM EGTA
1 M Urea
1% sodium deoxycholate
1% NP-40
0.1% SDS
Acknowledgments
We thank Stephanie Barker for technical assistance. We thank the following for financial support: Wellcome Trust Investigator Award WT210641/Z/18/Z to S.M.; O.B. acknowledges support from the Centre National de Recherche Scientifique, the Ecole Normale Superieure and the Institut National de la Sante et de la Recherche Medicale, France; N.F.I. acknowledges support from The Malaysia Ministry of Higher Education and IIUM Malaysia. The protocol is derived from Isa et al. (2021).
Competing interests
No competing interests.
References
- Chin, J. W. (2017). Expanding and reprogramming the genetic code. Nature 550(7674): 53-60.
- Hino, N., Okazaki, Y., Kobayashi, T., Hayashi, A., Sakamoto, K. and Yokoyama, S. (2005). Protein photo-cross-linking in mammalian cells by site-specific incorporation of a photoreactive amino acid. Nature Methods 2(3): 201-206.
- Isa, N. F., Bensaude, O., Aziz, N. C. and Murphy, S. (2021). HSV-1 ICP22 Is a Selective Viral Repressor of Cellular RNA Polymerase II-Mediated Transcription Elongation. Vaccines 9(10): 1054.
- Kobbi, L., Demey-Thomas, E., Braye, F., Proux, F., Kolesnikova, O., Vinh, J., Poterszman, A. and Bensaude, O. (2016). An evolutionary conserved Hexim1 peptide binds to the Cdk9 catalytic site to inhibit P-TEFb. Proc Natl Acad Sci U S A 113(45): 12721-12726.
- Neumann-Staubitz, P. and Neumann, H. (2016). The use of unnatural amino acids to study and engineer protein function. Curr Opin Struct Biol 38: 119-128.
- Wu, X., Spence, J. S., Das, T., Yuan, X., Chen, C., Zhang, Y., Li, Y., Sun, Y., Chandran, K., Hang, H. C. and Peng, T. (2020). Site-Specific Photo-Crosslinking Proteomics Reveal Regulation of IFITM3 Trafficking and Turnover by VCP/p97 ATPase. Cell Chem Biol 27(5): 571-585.e576.
- Xiao, H. and Schultz, P. G. (2016). At the Interface of Chemical and Biological Synthesis: An Expanded Genetic Code. Cold Spring Harb Perspect Biol 8(9): a023945.
- Ye, S., Köhrer, C., Huber, T., Kazmi, M., Sachdev, P., Yan, E. C. Y., Bhagat, A., RajBhandary, U. L. and Sakmar, T. P. (2008). Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. J Biol Chem 283(3): 1525-1533.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Isa, N. F., Bensaude, O. and Murphy, S. (2022). Amber Suppression Technology for Mapping Site-specific Viral-host Protein Interactions in Mammalian Cells. Bio-protocol 12(3): e4315. DOI: 10.21769/BioProtoc.4315.
Category
Drug Discovery > Drug Design
Microbiology > Microbe-host interactions > Virus
Biochemistry > Protein > Interaction > Crosslinking
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link