- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation of Healthy F4/80+ Macrophages from Embryonic day E13.5 Mouse Fetal Liver Using Magnetic Nanoparticles for Single Cell Sequencing
Published: Vol 11, Iss 23, Dec 5, 2021 DOI: 10.21769/BioProtoc.4243 Views: 3293
Reviewed by: Chiara AmbrogioEnrico PatruccoKarem A CourtZheng Zachory Wei

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Dan Wu [...] Weidong Wang
Oct 20, 2025 2118 Views
Selective Enrichment and Identification of Cerebrospinal Fluid-Contacting Neurons In Vitro via PKD2L1 Promoter-Driven Lentiviral System
Wei Tan [...] Qing Li
Nov 20, 2025 1260 Views
Revisiting Primary Microglia Isolation Protocol: An Improved Method for Microglia Extraction
Jianwei Li [...] Guohui Lu
Dec 5, 2025 1148 Views
Abstract
In vivo erythropoiesis occurs in the erythroblast island niche (EBI), comprising of a central macrophage that attaches to and aids the maturation of erythroid progenitors into mature reticulocytes. Macrophages in hematopoietic tissue such as embryonic fetal liver are heterogeneous and express the cell surface protein F4/80. Earlier methods of isolating F4/80+ macrophages from hematopoietic tissue relied on FACS sorting, but the relatively low numbers of F4/80+ cells obtained after FACS sometimes led to poor RNA quality. Additionally, since EBI macrophages are attached to erythroblasts, care must be taken to avoid contamination with bound erythroblasts. We have developed a novel method for isolating F4/80+ cells from E13.5 mouse fetal liver using magnetic nanoparticles, which can be performed on the lab bench. During cell suspension and homogenization, we also add a peptide that disrupts erythroid macrophage interactions and generates F4/80+ single cells free of erythroid contamination. Thus, our protocol generates a population enriched in F4/80+ cells that are healthy and ready for sensitive techniques such as single cell sequencing.
Background
Single cell sequencing technology has revolutionized our understanding of cell populations in a manner where cell types earlier perceived as homogeneous have been found to contain a great degree of heterogeneity. One such population comprises macrophages that support erythropoiesis in the erythroblast island (EBI) niche (Manwani and Bieker, 2008; Klei et al., 2017; Li et al., 2021). Previous studies have indicated that only a subpopulation of macrophages in erythroid tissues forms the EBI niche (Xue et al., 2014; Seu et al., 2017; Li et al., 2019), and single cell sequencing would be an excellent way to address the question of macrophage heterogeneity in erythropoietic tissue. However, since the EBI macrophage forms tight adhesive interactions with the maturing erythroblasts, removing erythroblasts from isolated macrophage populations is critical to the obtention of a pure population of macrophages for analysis.
Earlier efforts to isolate macrophages from hematopoietic tissues relied on using the macrophage-specific cell surface protein F4/80 as a marker for FACS-based methods (Xue et al., 2014; Li et al., 2019). However, macrophages comprise a relatively smaller fraction of the total cells in E13.5 fetal liver, and our previous experience with FACS sorting these cells showed that they resulted in poor quality of RNA or unhealthy cells that did not grow well in culture. Thus, for single cell sequencing, we decided to apply an isolation protocol using magnetic nanoparticles that usually generates healthier cells without compromising on purity (Figure 1). To reduce erythroid contamination in the final isolated F4/80+ macrophage population, during cell homogenization, we used a peptide that specifically inhibits the ICAM4/αv interactions that form the core of erythroblast-macrophage adhesive junctions (Xue et al., 2014). The EZ-Sep magnetic nanoparticle isolation kit from Cell Signaling Technologies utilizes a selection reagent that binds to phycoerythrin (PE) conjugated to a primary antibody such as anti-F4/80. The next step involves selection of F4/80-PE+ cells using the EZ-Sep PE selection cocktail (EZ-Sep mouse PE selection kit from Cell Signaling Technologies) and subsequent isolation using magnetic nanoparticles (Figure 1); this latter step can be repeated multiple times to increase purity. However, there is a trade-off where increased repetitions result in increased cell loss, which becomes a significant factor when dealing with fewer cell numbers. Thus, we repeated the purification step twice and were able to generate approximately 85% purity as assayed by flow cytometry for F4/80; in addition, the F4/80- cells were negative for erythroid markers indicating low erythroid contamination (Mukherjee et al., 2021).
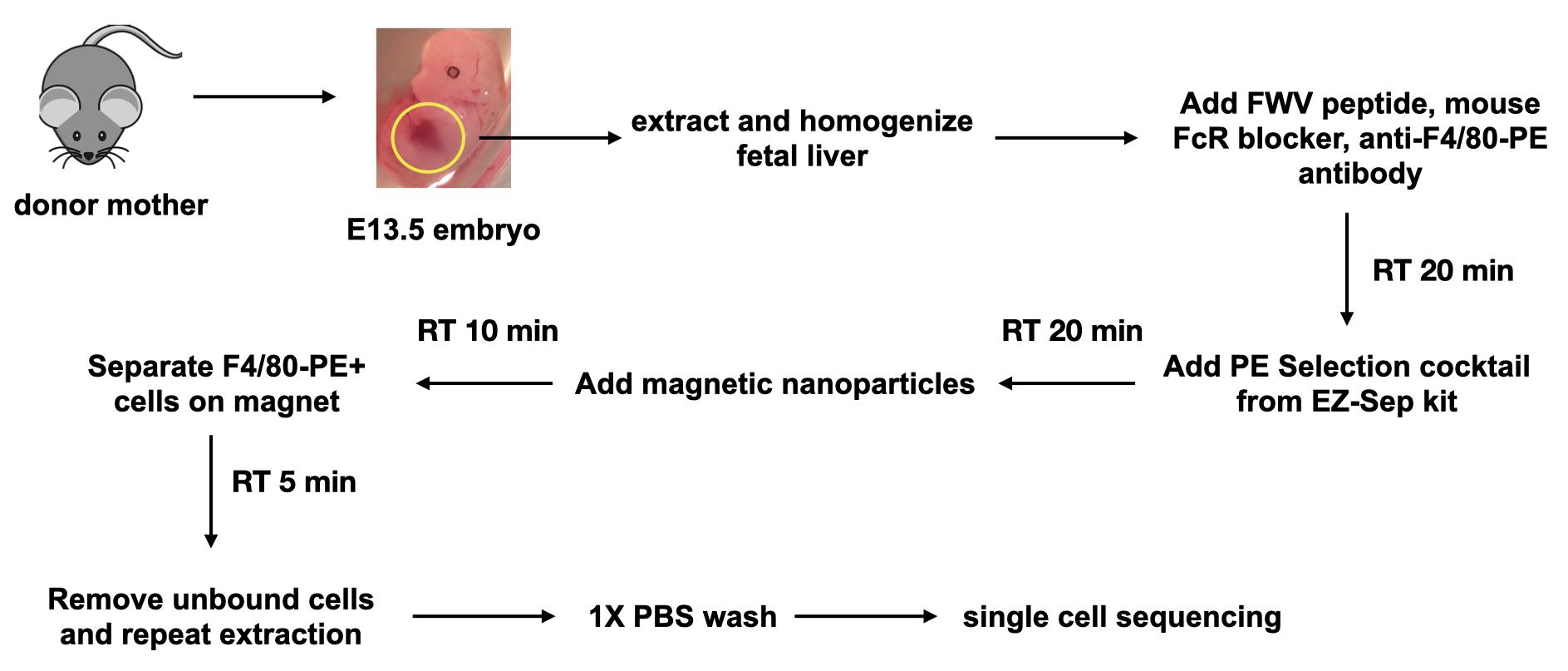
Figure 1. Schematic of F4/80-PE+ cell isolation for single cell sequencing using EZ-Sep magnetic nanoparticles.
Materials and Reagents
70 μm cell strainer (Fisher Scientific, Falcon, catalog number: 10788201)
Round bottom polystyrene 5 ml test tubes (Fisher Scientific, Falcon, catalog number: 14-959-5)
1.7 ml microfuge tubes (Crystalgen)
50 ml conical tubes (BD)
Sterile pipettes (Corning), tissue culture 6 cm dishes (BD)
Mouse E13.5 embryonic fetal liver
EasySep mouse PE positive selection kit (Cell Signaling Technologies, catalog number: 17656)
Anti-F4/80-PE antibody (rabbit polyclonal) (eBiosciences, catalog number: 12-4801-80)
FWV ICAM4/αv inhibitor peptide (custom sequence LRQGKTLRG) (Genscript, catalog number: SC1848). A stock solution was prepared at 23 mM and stored at -20°C. Stock solution is stable for at least 2 years.
Cell Culture Phosphate Buffered Saline 1× (Fisher Scientific, Corning, catalog number: MT21040)
Fetal Bovine Serum (GE Healthcare, HyClone, catalog number: SH30070.03)
0.5 M EDTA stock solution
Equipment
Pipettes (P200)
Dissection tools (e.g., sharp tweezers)
EasyEights EasySep Magnet (Cell Signaling Technologies, catalog number: 18103)
Refrigerated centrifuges capable of fitting 1.7 ml and 50 ml tubes
Flow Cytometer (Attune)
Procedure
Mouse E13.5 embryonic fetal liver extraction and cell homogenization
Sacrifice the mouse and extract E13.5 embryos, placing them in ice-cold 1× PBS in a sterile 6 cm tissue culture dish.
Using sharp tweezers, carefully extract the fetal livers into 200 μl of 1× PBS with 2% FBS.
Using a P200, pipette repeatedly but gently to homogenize the livers and generate single cell suspensions (collagenase or other enzymatic treatments are not required).
Filter the cells by passing them through a 70 μm mesh filter into 5 ml of ice-cold 1× PBS + 2% FBS in a 50 ml conical tube.
Mix well, take a small aliquot for counting the cells, and centrifuge the remaining cells in a refrigerated centrifuge at 180 × g for 5 min at 4°C.
F4/80-PE+ cell selection using EZ-Sep mouse PE selection kit
Resuspend the cells in 1× PBS + 2% FBS + 1 mM EDTA (EZ-Sep recommended medium) at room temperature and at a density of up to 108 cells per ml in a microfuge tube. (Usually, each fetal liver generates about 5-10 million cells, so each liver can be considered one sample). We usually resuspend each fetal liver in 200 µl of 1× PBS + 2% FBS in separate tubes.
Add FWV peptide to a final concentration of 2 mM and mix well (~17.5 µl of 23 mM stock solution added per 200 µl of sample).
Note: At this point onwards, the steps refer to the protocol for EZ-Sep mouse PE selection kit.
Add 2 µl of mouse FcR blocker from the EZ-Sep kit and pipette to mix cells.
At a dilution of 1:100 (based on the total volume from Step B1), add anti-F4/80-PE antibody, mix well by pipetting gently, and incubate at room temperature in a dark place for 20 min.
Note: Antibody dilutions will vary depending on the antibody used and must be standardized for different antibodies.
Add 2 µl of PE selection cocktail from the EZ-Sep kit, mix well by pipetting gently, and incubate at room temperature for 20 min.
Add 10 µl of magnetic nanoparticles from the EZ-Sep kit, mix well by pipetting gently, and incubate at room temperature for 10 min
Place the tubes on an EasyEights EasySep Magnet from Cell Signaling Technologies (which allows for eight tubes to be processed at once) for 5 min at room temperature.
Gently decant the supernatant by inverting the magnet with the tubes into a waste container (use one smooth inversion motion, do not shake).
Remove the tubes from the magnet and collect the cells in 1 ml of cold EZ-Seq recommended medium by gentle pipetting along the walls of the tubes. Centrifuge at 180 × g for 5 min at 4°C.
Resuspend in 200 µl of EZ-Sep recommended medium at room temperature, and repeat the magnetic nanoparticle selection steps from Step B6.
Remove the tubes from the magnet and wash twice in ice-cold 1× PBS. For each wash, add 2 ml of 1× PBS along the walls of the tubes and place on the magnet for 5min. Decant the liquid gently each time, as in Step B8.
After the final wash, resuspend the cells in 0.5 ml 1× PBS and proceed with single cell sequencing.
Take out a small aliquot of cells and analyze using flow cytometry for the purity of F4/80-PE+ cells.
Data analysis
Please refer to Mukherjee et al. (2021) for details of flow cytometry data analysis of purified F4/80+ population. In this study, 28,000 healthy F4/80+ macrophages (visually counted by light microscopy after Trypan Blue staining) were isolated from two independent wild-type E13.5 fetal livers and pooled for single cell sequencing. For details of single cell sequencing quality, cell heterogeneity, and data analysis, please refer to Mukherjee et al. (2021) . We obtained 13 independent clusters after U-MAP analysis using the Seurat package (Figure 2).
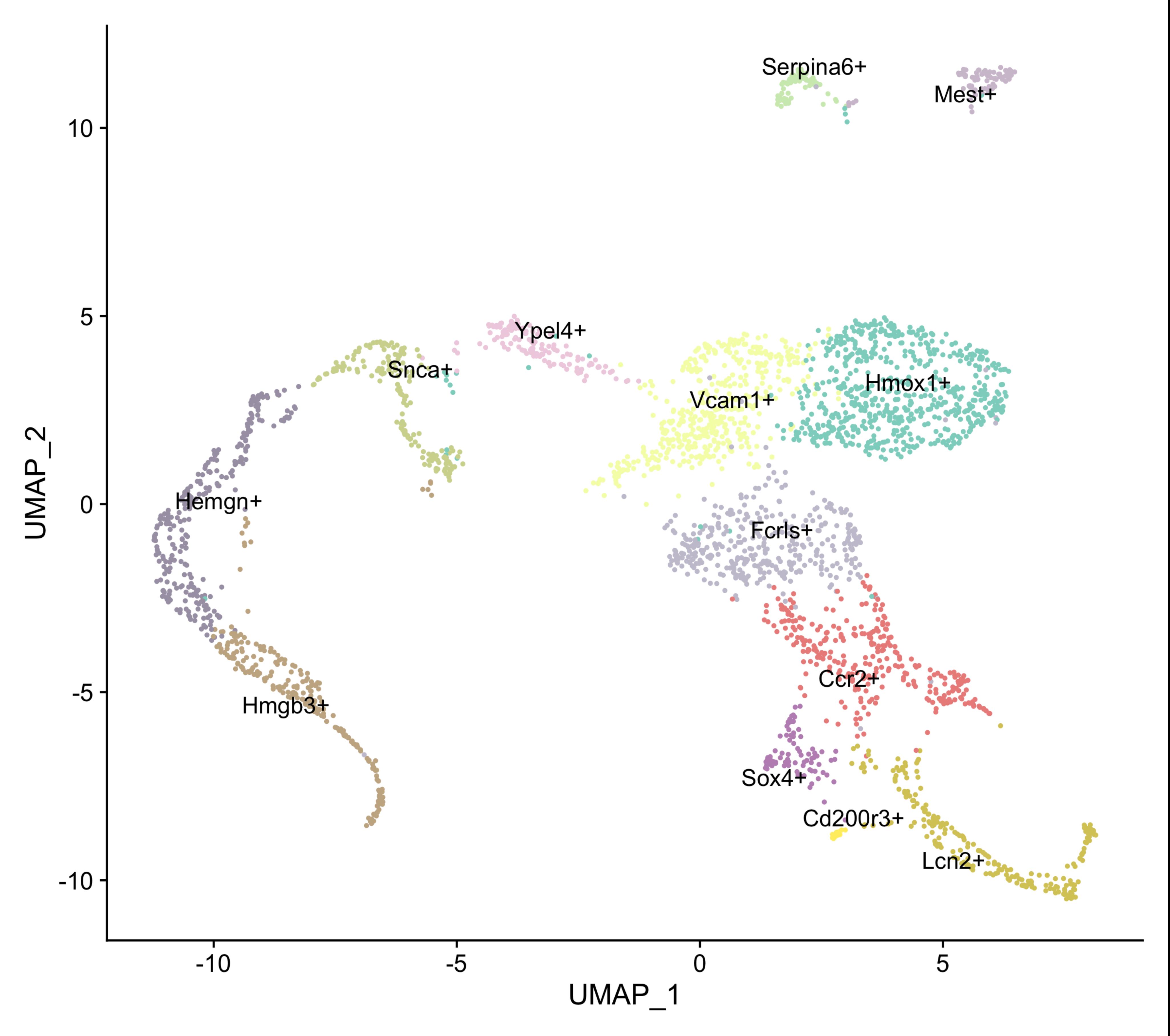
Figure 2. U-MAP clustering of single cell RNA-Seq data of F4/80+ macrophages isolated from E13.5 mouse fetal liver.
Acknowledgments
The authors would like to thank Xue Li for her input in designing an early version of this protocol. Flow cytometry was performed at the Sinai Flow Cytometry Core. This work was supported by NIH grants R01 DK102260 and DK121671 to JJB and by a Black Family Stem Cell Institute (ISMMS) Pilot Grant to KM.
This protocol was adapted with minor modification from previous study published by Mukherjee et al. (2021; Doi: 10.7554/eLife.61070).
Competing interests
The authors declare no competing interests
Ethics
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All animals were handled according to approved institutional animal care and use committee (IACUC) protocols (#18-1911) of the Mount Sinai School of Medicine.
References
- Klei, T. R., Meinderts, S. M., van den Berg, T. K. and van Bruggen, R. (2017). From the Cradle to the Grave: The Role of Macrophages in Erythropoiesis and Erythrophagocytosis. Front Immunol 8: 73.
- Li, W., Wang, Y., Zhao, H., Zhang, H., Xu, Y., Wang, S., Guo, X., Huang, Y., Zhang, S., Han, Y., et al. (2019). Identification and transcriptome analysis of erythroblastic island macrophages. Blood 134(5): 480-491.
- Li, W., Guo, R., Song, Y. and Jiang, Z. (2021). Erythroblastic Island Macrophages Shape Normal Erythropoiesis and Drive Associated Disorders in Erythroid Hematopoietic Diseases. Front Cell and Dev Biol 8: 613885.
- Manwani, D. and Bieker, J. J. (2008). The erythroblastic island. Curr Top Dev Biol 82: 23-53.
- Mukherjee, K., Xue, L., Planutis, A., Gnanapragasam, M. N., Chess, A. and Bieker, J. J. (2021). EKLF/KLF1 expression defines a unique macrophage subset during mouse erythropoiesis. Elife 10: e61070.
- Seu, K. G., Papoin, J., Fessler, R., Hom, J., Huang, G., Mohandas, N., Blanc, L. and Kalfa, T. A. (2017). Unraveling Macrophage Heterogeneity in Erythroblastic Islands. Front Immunol 8: 1140.
- Xue, L., Galdass, M., Gnanapragasam, M. N., Manwani, D. and Bieker, J. J. (2014). Extrinsic and intrinsic control by EKLF (KLF1) within a specialized erythroid niche. Development 141(11): 2245-2254.
Article Information
Copyright
Mukherjee and Bieker. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Mukherjee, K. and Bieker, J. J. (2021). Isolation of Healthy F4/80+ Macrophages from Embryonic day E13.5 Mouse Fetal Liver Using Magnetic Nanoparticles for Single Cell Sequencing. Bio-protocol 11(23): e4243. DOI: 10.21769/BioProtoc.4243.
- Mukherjee, K., Xue, L., Planutis, A., Gnanapragasam, M. N., Chess, A. and Bieker, J. J. (2021). EKLF/KLF1 expression defines a unique macrophage subset during mouse erythropoiesis. Elife 10: e61070.
Category
Cell Biology > Single cell analysis
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










