- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Oil Red O Staining for Lipid Content in Caenorhabditis elegans
Published: Vol 11, Iss 16, Aug 20, 2021 DOI: 10.21769/BioProtoc.4124 Views: 7617
Reviewed by: Demosthenis ChronisMichael EnosAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Lipidomic Analysis of Caenorhabditis elegans Embryos
Hung-Chi Yang [...] Daniel Tsun-Yee Chiu
Sep 20, 2017 11350 Views
Total Triglyceride Quantification in Caenorhabditis elegans
Anjali Sandhu and Varsha Singh
Nov 20, 2020 5211 Views

Rapid Lipid Quantification in Caenorhabditis elegans by Oil Red O and Nile Red Staining
Nicole L. Stuhr [...] Sean P. Curran
Mar 5, 2022 8373 Views
Abstract
The nematode Caenorhabditis elegans has emerged as a popular model system for studying the regulation of lipid metabolism. Therefore, it is critical to develop a method for determining fat storage in individual worms. Oil Red O (ORO) staining has been validated as an accurate assessment for major fat storage in C. elegans. Here, we describe an optimized protocol for ORO staining of C. elegans and provide detailed instructions for quantifying the intensity of ORO signal in images acquired by light microscopy.
Keywords: Oil Red OBackground
Recently, there has been a growing interest in studying the interplay between lipid metabolism and energy homeostasis. Caenorhabditis elegans has proved to be a powerful genetic system for elucidating complex physiological mechanisms and identifying novel genetic pathways. Lipid metabolism is a highly conserved process across eukaryotes. Many of the mammalian genes of the lipid metabolic pathways function similarly in C. elegans (Ashrafi, 2007; Jones and Ashrafi, 2009). Furthermore, several lipid regulatory pathways identified in C. elegans have been confirmed in mammalian systems (McKay et al., 2003; Suh et al., 2007). These features have made C. elegans an excellent animal model to study the regulation of fat accumulation and distribution.
To understand the molecular mechanisms regulating fat metabolism, a precise assessment of fat accumulation levels in worms is required. Various methods have been applied to visualize and measure fat content in C. elegans (Yen et al., 2010; Barros et al., 2012). Among them, Oil Red O (ORO) staining of fixed worms has been proved to be an accurate method to assess fat storage in C. elegans (O'Rourke et al., 2009; Zhang et al., 2010). ORO is a fat-soluble diazol dye that detects neutral lipids and cholesteryl esters but not biological membranes (Ramirez-Zacarias et al., 1992). Here, we demonstrate an ORO staining protocol with paraformaldehyde fixation. The method is readily applicable as it only requires standard laboratory and computer equipment. As an example of the application, we have included data on visualization and quantification of fat contents in wild-type N2, daf-2(e1370) mutants, and eat-2(ad1116) mutants.
Materials and Reagents
60 mm Petri dish (Greiner Bio-One, catalog number: 628102)
1.5 ml microcentrifuge tube (AXYGEN, catalog number: MCT-150-C)
Glass slide and coverslip (MARIENFELD, catalog number: 0107052)
10 ml syringe (Terumo, catalog number: DVR-3424)
0.22 μm syringe filters (MCE membrane filter, JET BIOFIL, catalog number: AGC-F-MCE022-13)
C. elegans strains were obtained from Caenorhabditis Genetics Center:
N2 Bristol, eat-2(ad1116), daf-2(e1370)
OP50-1 Escherichia coli bacteria (Caenorhabditis Genetics Center)
Oil Red O stain (Sigma, catalog number: O0625)
Sodium hypochlorite solution (6-14% active chlorine basis; Sigma, catalog number: 13440)
K2HPO4 (J.T. Baker, catalog number: 3252-01)
Bacto agar (BD, catalog number: 214510)
Bacteriological peptone (BD, catalog number: 211677)
CaCl2·2H2O (Sigma, catalog number: C3306)
Cholesterol (Sigma, catalog number: C3045)
EGTA (Sigma, catalog number: E8145)
Spermidine (Sigma, catalog number: S2626)
Spermine (Sigma, catalog number: S4264)
PIPES (Sigma, catalog number: P6757)
2-mercaptoethanol (Sigma, catalog number: M3148)
16% paraformaldehyde (Alfa Asear, catalog number: 43368)
KCl (J.T. Baker, catalog number: 3040-01)
2-Propanol (Sigma, catalog number: I9516)
KH2PO4 (Sigma, catalog number: P9791)
Na2HPO4·7H2O (OMNIPUR, catalog number: 8210)
NaCl (J.T. Baker, catalog number: 3624-05)
MgSO4·7H2O (Sigma, catalog number: M2773)
Triton X-100 (Sigma, catalog number: T8787)
LB broth (BIO-DOC, catalog number: BDLBB-500)
Nematode growth media (NGM) (see Recipes)
2× MRWB stock solution (see Recipes)
M9 buffer (see Recipes)
Nematode growth medium (NGM) agar for 1 L medium (see Recipes)
LB media (see Recipes)
Bleaching solution (see Recipes)
Oil Red O staining stock solution (see Recipes)
Potassium phosphate buffer (see Recipes)
Equipment
Low temperature refrigerated Incubator (Precision, catalog number: PR505755R, temperatures ranging from -10°C to +60°C)
Microcentrifuge (Thermo Scientific, model: Pico 17, 24 × 1.5/2 ml rotor, max. speed 13,300 rpm)
Dissecting microscope (Olympus, model: SZ61)
Fluorescence microscopy (Olympus, model: BX63)
Software
MatLab (The MathWorks, https://www.mathworks.com/products/matlab.html)
Cellsens (Olympus, https://www.olympus-lifescience.com/en/software/cellsens/)
Procedure
C. elegans maintenance
Inoculate an E. coli OP50 colony into 50 ml LB broth in a 200 ml flask and incubate with gentle agitation at 37°C overnight.
Apply approximately 200 μl of OP50 liquid culture to a 60 mm NGM plate. Incubate seeded plates at room temperature for 3-7 days before use.
Transfer 50-100 mixed-stage larval worms to a new OP50-seeded NGM plate using a heat-sterilized platinum wire pick. Keep worms at 20°C.
Repeat Step A3 every 2-3 days for maintenance.
C. elegans synchronization by bleaching
Collect worms (maintenance plates at Day 3 usually have the maxium numbers of gravid adults) in a 1.5 ml microcentrifuge tube with 1 ml of M9 buffer with 0.1% Triton X-100.
Centrifuge for 1 min at 200 × g and discard the supernatant.
Add 0.5 ml bleaching solution (see Recipe 4) into the tube and shake gently at room temperature for up to 5 min. Monitor the extent of worm lysis under the dissecting microscope.
Add 0.5 ml of M9 buffer to quench the bleaching process.
Note: Avoid over-bleaching.
Centrifuge for 1 min at 1,300 × g. Discard the supernatant.
Repeat Steps B4 and B5 twice to wash off the bleach solution.
Gently invert tubes and place two drops of 2-μl egg mixture on a clean microscope slide. Count eggs and determine the concentration of eggs.
Place ~150 eggs on an OP50 seeded 60 mm NGM plate (2-3 plates for each strain/condition) and grow them at 20°C.
Transfer synchronized worms to new OP50 plates every 2 days until they reach the desired stages.
Oil Red O staining of worms
Collect synchronized 300 worms (from two 60 mm NGM plates) at the desired stage with M9 + 0.1% Triton X-100 in a 1.5 ml centrifuge tube. Centrifuge for 1 min at 200 × g.
Discard the supernatant, then wash the worms with 1 ml of M9, and centrifuge for 1 min at 200 × g. Repeat this step three times to wash off bacteria. Leave worms in 100 μl M9 buffer after the last wash.
Prepare 1 ml of 2× MRWB buffer by adding 250 μl 16% paraformaldehyde to 750 μl MRWB stock solution.
Fix worms by adding 100 μl 2× MRWB buffer (freshly prepared) to the tube containing worms in 100 μl of M9 buffer.
Incubate samples at room temperature for 1 h. Gently invert the tubes every 15 min.
Spin down worms and discard the supernatant.
Add 1 ml of M9 + 0.1% Triton X-100, centrifuge for 1 min at 1,500 × g, and remove the supernatant. Repeat this step twice.
For dehydration, add 1 ml of 60% 2-propanol and then incubate samples for 15 min at room temperature in the dark.
While samples are under dehydration, prepare the Oil Red O working solution. Mix the Oil Red O stock solution (pre-heated in an 85°C water bath for 2 h) and ddH2O at a ratio of 3:2. Mix the working solution thoroughly. At this point, the mixed solution appears murky. The working solution should be made fresh from the stock solution each time.
Filter the mix solution through a 0.22-μm syringe filter. Effluent should appear dark red but clear.
Spin down dehydrated samples (from Step C8) at 1,500 × g for 1 min, and then discard the supernatant.
Add 200 μl of Oil Red O working solution (from Step C10) into each tube. Incubate in the dark at 37°C for 4-6 h.
Centrifuge at 1,500 × g for 1 min to pellet worms. Discard the staining dye.
Wash with 1 ml of M9 + 0.1% Triton X-100, and centrifuge for 1 min at 1,500 × g. Repeat this step twice.
After the last wash, leave 100 μl of the M9 solution. Store in the dark at room temperature until needed.
Mount the worms on a glass slide with an agarose pad (2% w/v agarose in ddH2O) and visualize them under the microscope.
Data analysis
The lipid contents of worms are quantified by measuring the area of staining and the average intensity of staining per worm. We have created a MatLab plugin for our image processing. Following are step by step instructions for our plugin (Figure 1):
Startup the MatLab software.
Go to HOME > Open > “ORO staining software”.
Go to Editor > Run. The Staining quality window will show up.
Press Open and select the images of the ORO staining worms. The Files window will also appear on the screen.
Press Set scale and adjust the blue line to match the scale bar. Enter the known distance, Pixel aspect ratio, and the Unit of length. Press OK to apply to selected images.
Set the threshold by adjusting the two horizontal bars in the Staining quantify window. The intensity values associated with the pixels in the red zone will be calculated. Click the image to see the original grayscale image.
Click ROI to select your region of interest in the image.
The average intensity of the red zone in a worm can be found in the Staining quantify window.
Click directly on the file name or use the Pre and Next button to select images.
After all measurements are done, press Save to save the results in Excel files.
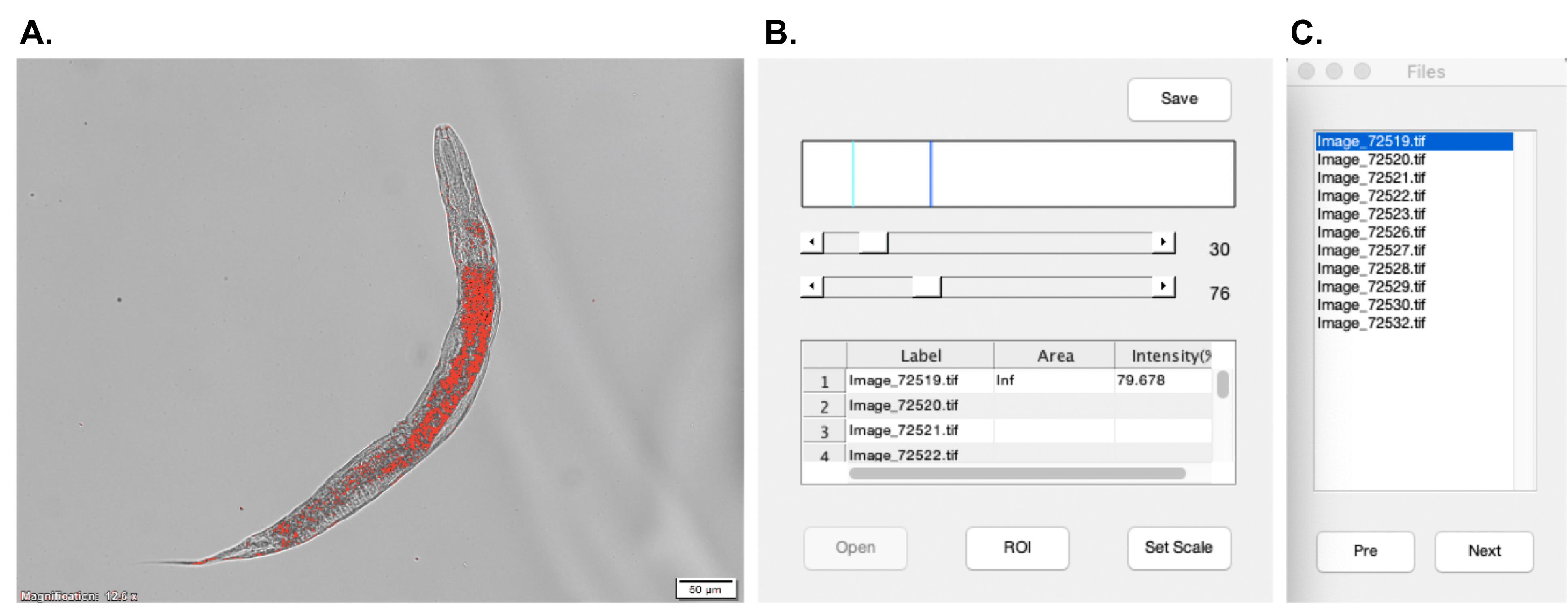
Figure 1. User interface of the ORO staining software. A. The "Image" window. The intensity values of pixels in the red area will be calculated as ORO signals. B. The "Staining quantify" window includes two threshold selection bars, the result chart, and the buttons for settings. C. The "Files" window. The images opened will be listed in this window.
In the following section, we have applied our ORO staining protocol to wild-type N2 animals, eat-2(ad1116) mutants, and daf-2(e1370) mutants. Previous studies have shown that eat-2(ad1116) mutants, a genetic model of dietary restriction, have less fat storage than wild-type animals (Avery, 1993). On the other hand, daf-2 mutants are long-lived and have increased fat content (Kimura et al., 1997). By following the protocol in the image processing and data analysis described above, we show that the area of ORO signals, but not their average intensity, was markedly decreased in eat-2 mutants. However, the area and intensity of ORO signals both increased in daf-2 mutants (Figure 2). Here, we have demonstrated that the results obtained through this protocol nicely align with previous findings and provide extra information on the total amount of fat content and the spatial distribution of stored fat in worms.

Figure 2. Oil Red O staining levels in wild-type N2, eat-2(ad1116), and daf-2(e1370) mutants. A. Representative images of Oil Red O staining in wild-type N2, eat-2(ad1116), and daf-2(e1370) animals at larval 4 stage (Scale bar: 50 μm). B. Quantification of the area stained by the Oil Red O in wild-type N2, eat-2(ad1116), and daf-2(e1370) mutants. C. Quantification of the average Oil Red O intensity in wild-type animals, eat-2(ad1116), and daf-2(e1370) mutants (100% describes the level of saturation). One-way ANOVA test followed by Dunnett's post-hoc test for selected groups was used. Sample size: n = 40 for N2, n = 45 for eat-2, n = 30 for daf-2. Results represent mean ± SD, and *** P < 0.001.
Notes
After fixation, worms get frequently stuck on plastic surfaces, such as tubes and pipets. To minimize the loss of worms, use glass droppers, glass tubes, and M9 buffer containing detergents, such as 0.1% Triton X-100. Nonetheless, a loss of 15-20% might occur throughout the process. Increase the number of worms if necessary.
When mounting the worms on slices, cut off the end of the pipette tips about 0.5 cm. Creating a broader opening will avoid breaking worms while transferring.
Recipes
M9 buffer
3 g KH2PO4
11.32 g Na2HPO4·7H2O
5 g NaCl
Water up to 1 L
Autoclave, then add 1 ml 1 M MgSO4 (filter to sterilize)
Nematode growth medium (NGM) agar for 1 L medium
3.0 g NaCl
20 g Bacto agar
2.5 g Bacteriological peptone
Autoclave to sterilize the agar, cool to 55°C, then add:
1 ml 1 M MgSO4 (filter to sterilize)
1 ml 1 M CaCl2 (filter to sterilize)
1 ml 5 mg/ml cholesterol in absolute ethanol
25 ml 1M Potassium phosphate buffer (see Recipe 6)
LB media
25 g LB broth dissolve in 1 L ddH2O
Bleaching solution
3 ml sodium hypochlorite solution
1.5 ml 5N KOH
Water up to 10 ml
MRWB stock solution
590 mg KCl
117 mg NaCl
0.7 ml of 1M EGTA
7.26 mg Spermidine
4 mg Spermine
1.5 ml of 1 M PIPES (pH 7.4)
0.1 ml β-ME
Disslove in 50 ml ddH2O
Oil Red O staining stock solution
0.5 g ORO powder in 100 ml of 2-propanol
Potassium phosphate buffer (1 M KPO4, pH 6.0)
108.3 g KH2PO4
35.6 g K2HPO4
Water up to 1 L
1 M EGTA
19 g EGTA powder in 50 ml of ddH2O
1 M HEPES Stock Solution (pH 7.4)
23.8 g of HEPES in 80 ml ddH2O
Add NaOH to raise the pH to 7.4
Water up to 100 ml
Acknowledgments
We thank Cho-Lun Chiang for providing technical support for the data analysis. This work was supported by grants from the Ministry of Science and Technology, Taiwan (MOST 106-2311-B-010-005). The protocol is based on the publication “S-adenosyl methionine synthetase SAMS-5 mediates dietary restriction-induced longevity in Caenorhabditis elegans” (Chen et al., 2020).
Competing interests
The authors have no conflicts of interest to declare.
References
- Ashrafi, K. (2007). Obesity and the regulation of fat metabolism. WormBook 1-20.
- Avery, L. (1993). The genetics of feeding in Caenorhabditis elegans. Genetics 133(4): 897-917.
- Barros, A. G., Liu, J., Lemieux, G. A., Mullaney, B. C. and Ashrafi, K. (2012). Analyses of C. elegans fat metabolic pathways. Methods Cell Biol 107: 383-407.
- Chen, C. C., Lim, C. Y., Lee, P. J., Hsu, A. L. and Ching, T. T. (2020). S-adenosyl methionine synthetase SAMS-5 mediates dietary restriction-induced longevity in Caenorhabditis elegans. PLoS One 15(11): e0241455.
- Jones, K. T. and Ashrafi, K. (2009). Caenorhabditis elegans as an emerging model for studying the basic biology of obesity. Dis Model Mech 2(5-6): 224-229.
- Kimura, K. D., Tissenbaum, H. A., Liu, Y. and Ruvkun, G. (1997). daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277(5328): 942-946.
- McKay, R. M., McKay, J. P., Avery, L. and Graff, J. M. (2003). C. elegans: a model for exploringthe genetics of fat storage. Dev Cell 4(1): 131-142.
- O'Rourke, E. J., Soukas, A. A., Carr, C. E. and Ruvkun, G. (2009). C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab 10(5): 430-435.
- Ramirez-Zacarias, J. L., Castro-Munozledo, F. and Kuri-Harcuch, W. (1992). Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil red O. Histochemistry 97(6): 493-497.
- Suh, J. M., Zeve, D., McKay, R., Seo, J., Salo, Z., Li, R., Wang, M. and Graff, J. M. (2007). Adipose is a conserved dosage-sensitive antiobesity gene. Cell Metab 6(3): 195-207.
- Yen, K., Le, T. T., Bansal, A., Narasimhan, S. D., Cheng, J. X. and Tissenbaum, H. A. (2010). A comparative study of fat storage quantitation in nematode Caenorhabditis elegans using label and label-free methods. PLoS One 5(9).
- Zhang, S. O., Trimble, R., Guo, F. and Mak, H. Y. (2010). Lipid droplets as ubiquitous fat storage organelles in C. elegans. BMC Cell Biol 11: 96.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wang, F. and Ching, T. (2021). Oil Red O Staining for Lipid Content in Caenorhabditis elegans. Bio-protocol 11(16): e4124. DOI: 10.21769/BioProtoc.4124.
Category
Biochemistry > Lipid > Lipid measurement
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









