- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Assay for Assessing Mucin Binding to Bacteria and Bacterial Proteins
(*contributed equally to this work) Published: Vol 11, Iss 5, Mar 5, 2021 DOI: 10.21769/BioProtoc.3933 Views: 4763
Reviewed by: Emilia KrypotouElizabeth LibbyAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Producing GST-Cbx7 Fusion Proteins from Escherichia coli
Thao Ngoc Huynh and Xiaojun Ren
Jun 20, 2017 11523 Views
Combining Gel Retardation and Footprinting to Determine Protein-DNA Interactions of Specific and/or Less Stable Complexes
Meng-Lun Hsieh [...] Deborah M. Hinton
Dec 5, 2020 3581 Views
Large-scale Purification of Type III Toxin-antitoxin Ribonucleoprotein Complex and its Components from Escherichia coli for Biophysical Studies
Parthasarathy Manikandan [...] Mahavir Singh
Jul 5, 2023 2169 Views
Abstract
Legionella pneumophila, a Gram-negative bacterium and the causative agent of Legionnaires’ disease, exports over 300 effector proteins/virulence factors, through its type II (T2SS) and type IV secretion systems (T4SS). One such T2SS virulence factor, ChiA, not only functions as a chitinase, but also as a novel mucinase, which we believe aids ChiA-dependent virulence during lung infection. Previously published protocols manipulated wild-type L. pneumophila strain 130b and its chiA mutant to express plasmid-encoded GFP. Similarly, earlier studies demonstrated that wheat germ agglutinin (WGA) can be fluorescently labeled and can bind to mucins. In the current protocol, GFP-labeled bacteria were incubated with type II and type III porcine stomach mucins, which were then labeled with TexasRed-tagged WGA and analyzed by flow-cytometry to measure the binding of bacteria to mucins in the presence or absence of endogenous ChiA. In addition, we analysed binding of purified ChiA to type II and type III porcine stomach mucins. This protocol couples both bacterial and direct protein binding to mucins and is the first to measure Gram-negative bacterial binding to mucins using WGA and flow-cytometric analysis.
Graphic abstract:
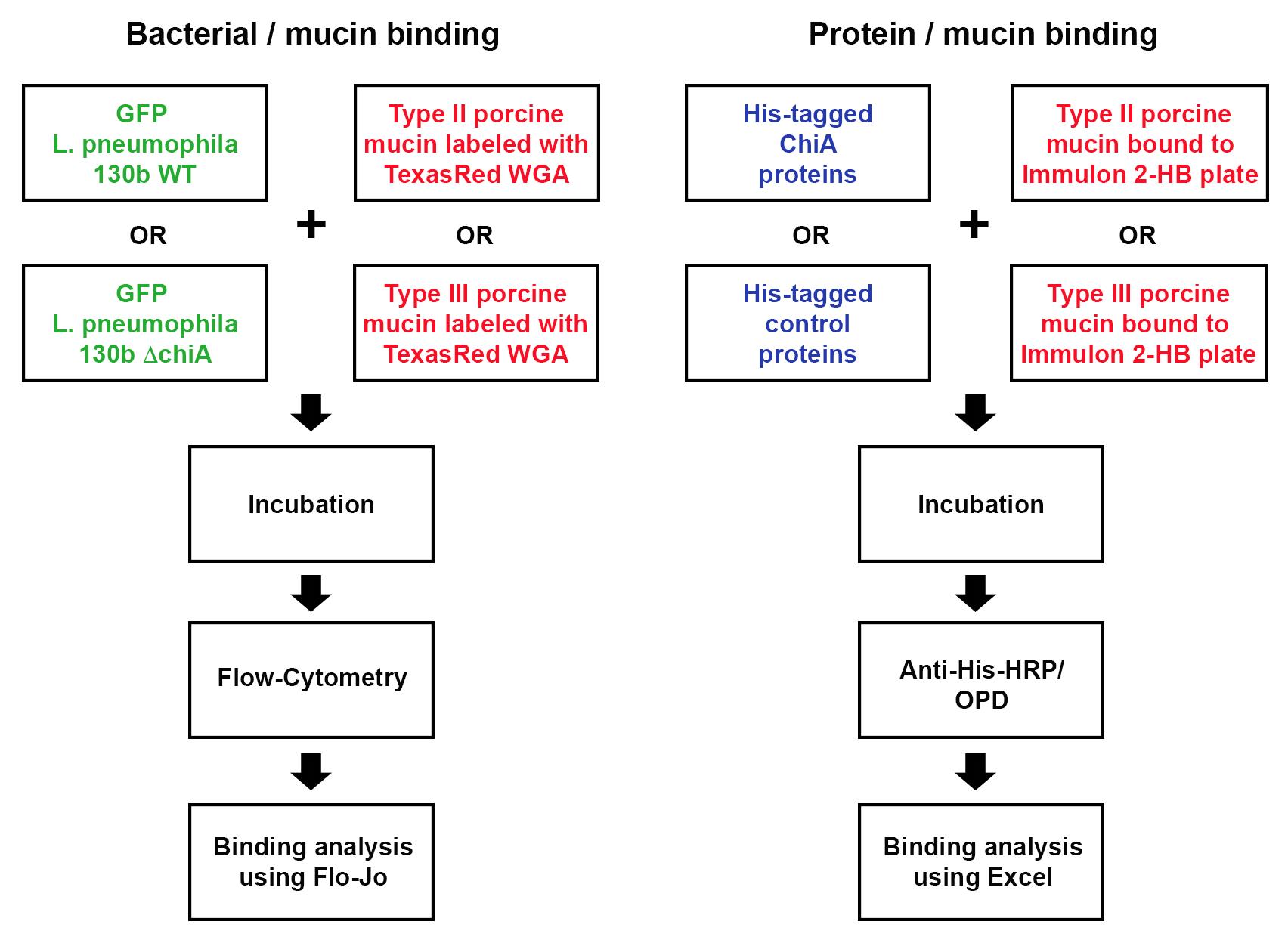
Strategy for assessing bacterial and protein binding to mucins
Background
Legionella pneumophila (Lpn), a Gram-negative bacterium, is the causative agent of Legionnaires’ disease, a severe form of pneumonia. Lpn is an intracellular pathogen that produces over 300 protein effectors that it secretes through either a type II secretion system (T2SS), or through a type IV secretion system (T4SS) (Hubber and Roy, 2010; White and Cianciotto, 2019). ChiA is one such T2SS protein effector. ChiA is an 81-kDa endochitinase that has a role in Lpn virulence during lung infection (Rehman et al., 2020). Lpn carrying a deletion of the ChiA gene (ΔchiA) shows decreased survival in the lungs of mice, compared to WT Lpn (DebRoy et al., 2006). While humans do not produce chitin, they do produce analogous glycoproteins, mucins, that have known properties in interacting with and blocking infection of other pathogens. In our study, we showed, for the first time, that ChiA is able to both bind to and degrade mucins (Rehman et al., 2020). To determine if live bacteria were able to bind to mucins, we utilized our previously published protocol to manipulate wild-type Lpn strain 130b and its chiA mutant to express a GFP-producing plasmid (DebRoy et al., 2006; Rondelet and Condemine, 2013). Furthermore, earlier studies showed that wheat germ agglutinin (WGA) binds to mucins (Bhavanandan and Katlic, 1979; Valdizan et al., 1992). Therefore, we utilized porcine stomach mucins which we labeled with TexasRed-conjugated WGA (Model et al., 2009). To determine whether ChiA directly binds to these mucins, we also used an ELISA based-assay with purified recombinant (N-terminally His-tagged) ChiA and detected binding using anti-His6 antibodies.
Although mucin binding to bacteria has been studied previously (Naughton et al., 2014), and fluorescently labeled WGA has been studied in the context of Gram-positive bacteria (Fife et al., 2000) this is the first protocol to directly label mucins and bacteria and then utilize flow cytometry to measure mucin binding to a Gram-negative bacterium. Furthermore, by analyzing both bacterial binding to mucins and the binding of purified proteins to mucins, this protocol provides insight into synergistic binding of different surface exposed bacterial proteins to mucins. This protocol can be further applied to study mucin binding to other Gram-negative bacteria.
Part I: Bacterial Mucin Binding with Flow Cytometry
Materials and Reagents
Legionella pneumophila Brenner et al. (1979) (ATCC® BAA-74TM)
WT 130b purchased from ATCC (see above)
ChiA mutant as previously reported (DebRoy et al., 2006)
GFP plasmid (pMGFP), as previously reported, was derived from pMMB-GRN/pMMB-Gent (addgene 45475). GFP is expressed from a Ptac promotor and therefore IPTG is required (White and Cianciotto, 2016) (Sturgill-Koszycki and Swanson, 2000). While copy number in Legionella is unknown, the PMMB-Gent derives from PMMB67EH (ATCC 37622)
1× Phosphate Buffered Saline (PBS) (Corning, catalog number: 21-040-CM )
IPTG (Sigma-Aldrich, catalog number: I6758 )
Type II porcine stomach mucin (Sigma-Aldrich, catalog number: M1778 )
Type III porcine stomach mucin (Sigma-Aldrich, catalog number: M2378 )
TexasRed-tagged WGA (ThermoFisher Scientific, catalog number: W21405 )
Sodium carbonate, Na2CO3 (Sigma-Aldrich, catalog number: S7795 )
Sodium bicarbonate, NaHCO3 (Sigma-Aldrich, catalog number: S5761 )
Buffered Charcoal Yeast Extract (BCYE) agar plates (see Recipes)
ACES (Sigma-Aldrich, catalog number: A9758 )
KOH (Sigma-Aldrich, catalog number: 221473 )
Yeast Extract (Sigma-Aldrich, catalog number: Y1625 )
α-Ketoglutaric acid sodium salt (Sigma-Aldrich, catalog number: K1875 )
Activated Charcoal (Sigma-Aldrich, catalog number: C9157 )
Bacteriological Agar (Sigma-Aldrich, catalog number: A5306 )
L-cysteine HCl (Sigma-Aldrich, catalog number: C1276 )
Ferric pyrophosphate (Sigma-Aldrich, catalog number: P6526 )
50 mM Carb/Bicarb Buffer (see Recipes)
Mucin solution (see Recipes)
Equipment
Flow Cytometer (https://www.bdbiosciences.com/en-us/go-campaign/lsr-ii-comp-cont) using Blue Laser (488 nm), Long pass Filter 600 and 505, Band Pass Filter 600-620 and 500-550 (BD Biosciences, model: LSR II )
General Purpose UV/Vis Spectrophotometer (Beckman Coulter, model: DU720 )
Forced Air Microbiological Incubators (VWR, catalog number: 89511-430 )
Software
GraphPad Prism version 8.0.0 for Mac (GraphPad Software, San Diego, California USA, www.graphpad.com)
FlowJoTM Software for Mac, Version 8.8.6. (Ashland, OR: Becton, Dickinson and Company, www.flowjo.com)
Procedure
Lpn Preparation
Streak out Lpn strain 130b and ChiA mutant NU319, both harboring a GFP-expressing plasmid, onto BCYE agar plates containing IPTG at 1 mM.
Incubate the plates for 3 days in a 37 °C incubator to grow out a lawn of bacteria.
Resuspend ~3 swabs of scraped bacteria into 5 ml of PBS and read the optical density (OD) of each suspension at 660 nm using a DU720 spectrophotometer. Use PBS to dilute the bacterial suspension to an OD of 0.3, which corresponds to approx. 1 × 109 CFU/ml.
Mucin Preparation
Prepare both type II and type III porcine stomach mucin solutions in carb/bicarb buffer according to the recipe below.
Mucin/Lpn Incubation
Statically incubate 1 ml of the Lpn suspension with 100 μl of either type II porcine stomach mucin, type III porcine stomach mucins, or PBS for 1 h in a 25 °C or 37 °C incubator. Repeat every condition in triplicate.
Note: Keep an aliquot of the type II and type III porcine stomach mucin solutions (no bacteria) as controls for the flow-cytometry analysis.
Centrifuge, at room temperature, the mixtures for 5 min at 4,000 × g (for the 37 °C samples) or 8,000 × g (for the 25 °C samples).
Wash each pellet in 1 ml of PBS
Repeat centrifugation and wash step (Steps C2 and C3) three times.
Resuspend pellet in 1 ml of PBS containing 7.5 μg of TexasRed-tagged WGA and incubate statically for 15 min at 25 °C.
Repeat the centrifugation and wash steps (Steps C2 and C3) three times.
Resuspend the pellet in 500 µl of PBS.
Flow Cytometry
Use BD LSRII flow cytometer to analyze suspensions via a TexasRed filter (Blue Laser – 488 nm: Long Pass 600, Band Pass 600-620) and GFP Filter (Blue Laser – 488 nm: Long Pass 505, Band Pass 500-550).
Set gating TexasRed and GFP parameters using only mucin and only bacteria respectively.
Collect a minimum of 500,000 events.
Data analysis
The experiment was replicated a minimum of three times, and each experiment had three technical replicates. FlowJo software was used to analyze percent of mucin binding to Lpn. Bacteria capable of expressing GFP and isolated mucin were used to set thresholds for both the TexasRed- and GFP-negative populations (unlabeled mucins and no IPTG-dependent GFP expression) and the TexasRed- and GFP-positive populations (TexasRed labeled mucins and IPTG-induced GFP expression). After setting these thresholds (see below for the four histograms on the left), the percent population (see below for quadrant analysis in the right-most flow-plot) of single GFP-positive (Quadrant [Q]3), single TexasRed-positive (Q1), and double GFP and TexasRed positive (Q2) were calculated using FloJo quadrant tool. Thus, Q2 was used as a calculation of percent of mucin binding to bacteria. Due to variability in mean fluorescence between experimental days, data were normalized to GFP-Lpn incubated with TexasRed fluorophore alone (background), see sample data table below and Figure 4D (Rehman et al., 2020). GraphPad Prism8 statistical software was used to graph and analyze the percent binding of bacteria to mucins. Prism8 analysis quantified standard deviation and used the two-tailed Student’s t-test function to test for significant differences between groups tested. Only P-values less then 0.05 were determined to be significant.
Flow Cytometry Protocol (Figure 1)
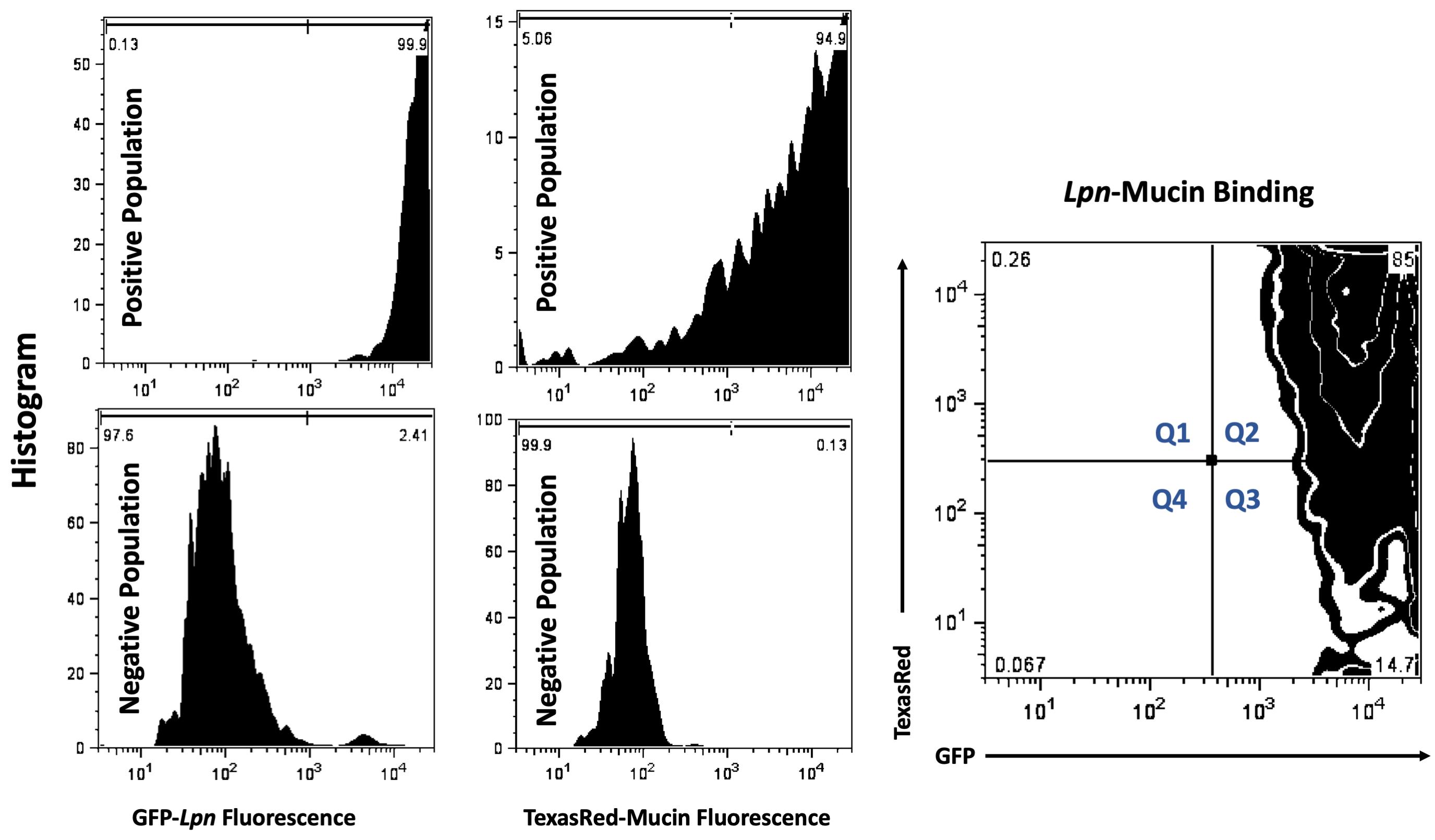
Figure 1. Flow Cytometry. Thresholds were set using single-positive samples 1. IPTG induced GFP production in Lpn (top left histogram) and 2. non-GFP bacteria with TexasRed labeled mucins (top middle histogram). Single negative populations were set using 1. Bacteria cultured without IPTG (bottom left histogram) 2. Non-GFP bacteria with un-labeled mucins (bottom middle histogram). For sample analysis, IPTG induced, GFP-expressing bacteria were co-incubated with TexasRed-labeled mucins. Gating parameters were used to setup quadrant percentiles for analysis. All events collected were partitioned into four quadrants using FloJo tool. Percentage of total population that were only TexasRed positive were labeled Q1, double GFP and TexasRed bacterial populations were labeled Q2, single GFP-positive populations were designated Q3 and non-fluorescent, all negative, populations were designated Q4. Percent of population that was GFP and TexasRed double positive (Q2) was used as a metric for mucin-bacteria binding and was analyzed in the sample table (Table 1) below.
Sample Data (Table 1)
Table 1. Percent of population that was double positive for TexasRed-mucin and GFP-Lpn (Q2) was used as a value for mucin binding to bacteria. In the single example of WT behavior in the flow plot shown above, this value was 85% (right-most plot in Figure 1). This value plus the values obtained from two additional independent replicates testing WT bacteria were added to the third row in the first three columns on the left, i.e., under headers Rep1, Rep2, Rep3. The raw values for three technical replicates obtained from analysis of another bacterial sample; i.e., a ChiA mutant, appear in the three left-most lanes in row-4. The value for each of the replicates was then normalized to GFP-bacteria (WT or mutant) incubated with TexasRed alone (as noted in rows 1 and 2), to quantify background TexasRed fluorophore binding to bacteria. For the two bacterial samples incubated with mucin, these values appear in columns 5, 6, and 7 for rows 3 and 4. Finally, the normalized values were averaged, and standard deviations calculated, as indicated in the last two columns. In this sample dataset, 22.5 + 5.4% of WT bacteria significantly bound the mucin.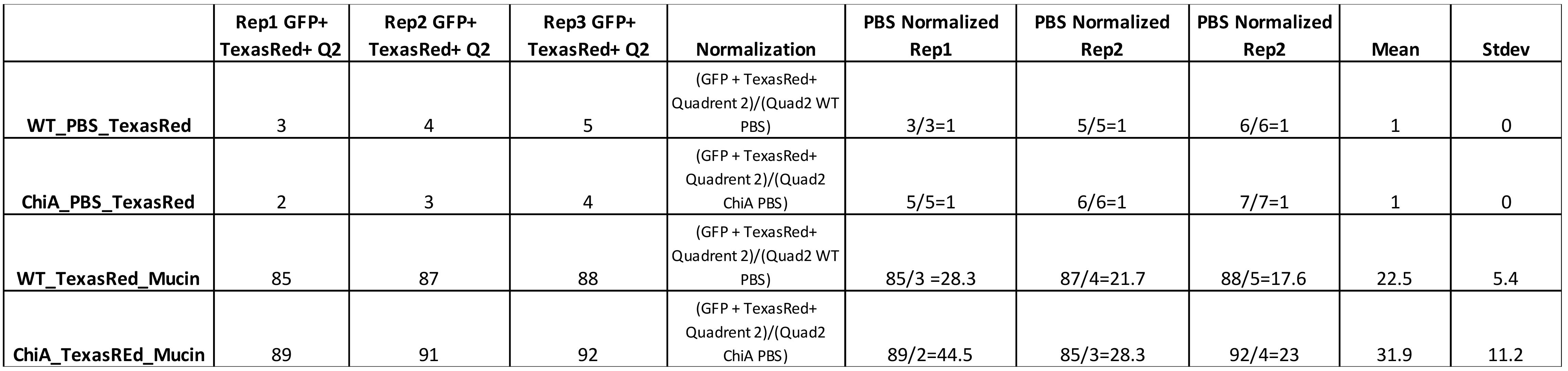
Recipes
BCYE agar plates
Add 10 g of ACES to ~900 ml of double distilled water. Stir into solution
Add ~2.2 g KOH. Stir into solution
Add 10 g of Yeast Extract and 1 g α-ketoglutaric acid. Stir into solution
Adjust the pH of the broth to 6.85-6.95. Use concentrated HCl or 10 N KOH as required. Add double distilled water to 1 L
Dispense the broth into a 2-L flask
Add 1.5 g activated charcoal and 15 g of agar
Autoclave 20 min at 121 °C
Prepare cysteine and iron solutions respectively by adding 0.4 g of L-cysteine HCl to 10 ml double distilled water and 0.25 g ferric pyrophosphate to 10 ml double distilled water. Once each solution is made, sterilize the solution by filter (0.22 µm).
Cool the medium to 50 °C in a water bath after autoclaving
Add separately 10 ml filter-sterilized (0.22 µm) cysteine and 10 ml filter-sterilized (0.22 µm) ferric pyrophosphate to the medium
If making plates with IPTG, add 1 ml of 1 M Filter Sterilized (0.22 µm) IPTG
Cool to 40 °CPour plates and store at 4 °C
once solidified
- 50 mM Carb/Bicarb Buffer
Dissolve 1.59 g Na2CO3 and 2.93 g NaHCO3 in 1 L of deionized water
Adjust the pH to 9.6 using HCl or NaOH
- Mucin solution
Stir for 30 min to dissolve 10 mg of either type II or type III porcine stomach mucin in 100 ml of 50 mM Carb/Bicarb buffer above. Autoclave for 15 min at 135 °C to sterilize and to further dissolve the mucin*.
*Note: Mucin is difficult to dissolve and will not fully go into solution by mixing alone; thus, autoclaving is required to get a homogeneous solution. Fresh preparation of mucin is recommended for each experiment.
Part II: Recombinant Protein Mucin Binding with ELISA
Materials and Reagents
Immulon 2-HB 96-well plates (VWR, catalog number: 735-0464)
Purified N-terminally His-tagged protein samples
Anti-His HRP-conjugated antibody (Sigma-Aldrich, catalog number: SAB4301134)
Mucins from porcine stomachs type II (Sigma-Aldrich, catalog number: M2378 )
Mucins from porcine stomachs type III (Sigma-Aldrich, catalog number: M1778 )
Sodium carbonate, Na2CO3(Sigma-Aldrich, catalog number: S7795 )
Sodium bicarbonate, NaHCO3(Sigma-Aldrich, catalog number: S5761 )
Bovine serum albumin (BSA) (Melford, catalog number: A30075) Phosphate buffered saline (PBS) (Fisher Scientific, catalog number: BP399-1)
Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E9884)Zinc chloride, ZnCl2(Sigma-Aldrich, catalog number: 229997)
Tween 20 (Sigma-Aldrich, catalog number: P1379)
o-Phenylenediamine dihydrochloride tablets (OPD) (Sigma-Aldrich, catalog number: P9187)
SnakeSkin® dialysis tubing (Thermo Scientific, catalog number: 68100)
Incubation buffer (see Recipes)
50 mM Carb/Bicarb Buffer (see Recipes)
Mucin solution (see Recipes)Blocking buffer (see Recipes)
Equipment
Plate reader (LabSystems iEMS Reader MF, catalog number: 5921200) using 450 nm filter
Incubator (New Brunswick Innova 4230)
Centrifuge (Eppendorf, model: 5810R)
Software
Ascent Microplate Reader Software version 2.7.0 for Windows
Microsoft Excel version 16.43 for Mac; although any database analysis software is appropriate
Procedure
Protein preparation
Dialyze sample proteins at 100 μM into incubation buffer, prepared according to the recipe below. Adjust the final protein concentration after dialysis to 10 μM.Mucin preparation
Prepare both type II and type III porcine stomach mucin solutions in carb/bicarb buffer according to the recipe below.Incubation
Statically incubate an Immulon 2-HB 96-well plate with 50 μl of either type II porcine stomach mucin, type III porcine stomach mucins, or PBS overnight in a 4 °C incubator. Repeat every condition in triplicate.
Remove the mucin solution and block the wells with 200 μl of blocking buffer, according to the recipe below, statically for 1 h in a 24 °C incubator.
Remove the blocking buffer and wash once by adding 200 μl of incubation buffer and incubating statically on the bench for 5 min. Next incubate the wells statically for 3 h in a 24 °C incubator with 50 μl of purified protein samples or PBS.
Wash with 200 μl of incubation buffer as described above and repeat for a total number of 4 washing steps.
- Detection
Remove the buffer and incubate the wells with 50 μl of anti-His-HRP antibody, diluted 1:2,000 in incubation buffer, statically for 1 h in a 24 °C incubator.
Wash four times as described above with 200 μl of incubation buffer.
Add 150 μl of o-Phenylenediamine dihydrochloride to each well and leave the plate in a 24 °C incubator without shaking for 30 min in the dark.
Record at 450 nm using a plate reader.
Data analysis
The experiment was replicated a minimum of three times, and each experiment had three technical replicates. Data were imported into Microsoft Excel software and baseline corrected with the incubation with PBS alone samples. Sample data for ChiA, NttE (negative control) and SslE (positive control) is given below for binding to type II porcine stomach mucin (also see Figure 4C, Rehman et al., 2020). Excel analysis provided standard deviation values and a two-tailed Student’s t-test function was used to test for significant differences between groups tested. Only P-values less then 0.05 were determined to be significant.
Sample Data (Table 2)
Table 2. Example data for proten binding to type II stomach mucin extracts
| Rep1 | Rep2 | Rep3 | Mean (PBS) | PBS subtracted Rep 1 | PBS subtracted Rep 2 | PBS subtracted Rep 3 | Mean (mucin) | Stdev (mucin) | |
| ChiA_PBS | 0.239 | 0.190 | 0.181 | 0.203 | - | - | - | - | - |
| NttE_PBS (-) | 0.084 | 0.074 | 0.062 | 0.073 | - | - | - | - | - |
| SslE_PBS (+) | 0.130 | 0.164 | 0.156 | 0.150 | - | - | - | - | - |
| ChiA_mucin | 0.933 | 0.846 | 0.900 | - | 0.730 | 0.643 | 0.697 | 0.690 | 0.044 |
| NttE_mucin (-) | 0.138 | 0.119 | 0.151 | - | 0.065 | 0.046 | 0.078 | 0.063 | 0.016 |
| SslE_mucin (+) | 0.718 | 0.912 | 0.910 | - | 0.568 | 0.762 | 0.760 | 0.697 | 0.111 |
Recipes
Incubation buffer (prepare fresh and store at 4 °C)
Mix/dissolve 0.5 g BSA and 0.5 ml Tween-20 for every 1 L of PBS buffer
Cool to 4 °C
- 50 mM Carb/Bicarb Buffer (can be prepared in advance and stored at room temperature)
Dissolve 1.59 g Na2CO3 and 2.93 g NaHCO3 in 1 L of deionized water
Adjust the pH to 9.6 using HCl or NaOH
- Mucin solution (can be prepared the day before and stored at 4 °C)
Stir for 30 min to dissolve 10 mg of either type II or type III porcine stomach mucin in 100 ml of 50 mM Carb/Bicarb buffer above.
Autoclave for 15 min at 135 °C to sterilize and to further dissolve the mucin*.
*Note: Mucin is difficult to dissolve and will not fully go into solution by mixing alone; thus, autoclaving is required to get a homogeneous solution.
Blocking buffer (can be prepared the day before and stored at 4 °C)
Mix/dissolve 0.1 g BSA and 0.5 ml Tween-20 for every 1 L of PBS buffer
Cool to 4 °C
Acknowledgments
Work in the Cianciotto lab was supported by a National Institutes of Health grant R01AI 043987. LSG and RCW were also partly supported by National Institutes of Health training grants T32 GM08061 and T32 AI0007476, respectively. We thank the Northwestern ImmunoBiology Flow Cytometry Core for the maintenance and use of their equipment. Work in the Garnett lab was supported by MRC grants MR/M009920/1 and MR/R017662/1.
Competing interests
NPC and JAG have a pending patent application (63/005,592) that describes the use of ChiA for therapeutic applications.
Ethics
No human or animal subjects were used in this protocol.
References
Bhavanandan, V. P. and Katlic, A. W. (1979). The interaction of wheat germ agglutinin with sialoglycoproteins. The role of sialic acid. J Biol Chem 254(10): 4000-4008.
- Brenner, D. J., Steigerwalt, A. G. and McDade, J. E. (1979). Classification of the Legionnaires' disease bacterium: Legionella pneumophila, genus novum, species nova, of the family Legionellaceae, familia nova. Ann Intern Med 90(4): 656-658.
- DebRoy, S., Dao, J., Soderberg, M., Rossier, O. and Cianciotto, N. P. (2006). Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc Natl Acad Sci U S A 103(50): 19146-19151.
- Fife, D. J., Bruhn, D. F., Miller, K. S. and Stoner, D. L. (2000). Evaluation of a fluorescent lectin-based staining technique for some acidophilic mining bacteria.Appl Environ Microbiol 66(5): 2208-2210.
- Hubber, A. and Roy, C. R. (2010). Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol 26: 261-283.
- Model, M. A., Reese, J. L. and Fraizer, G. C. (2009). Measurement of wheat germ agglutinin binding with a fluorescence microscope. Cytometry A 75(10): 874-881.
- Naughton, J., Duggan, G., Bourke, B. and Clyne, M. (2014). Interaction of microbes with mucus and mucins: recent developments. Gut Microbes 5(1): 48-52.
- Rehman, S., Grigoryeva, L. S., Richardson, K. H., Corsini, P., White, R. C., Shaw, R., Portlock, T. J., Dorgan, B., Zanjani, Z. S., Fornili, A., Cianciotto, N. P. and Garnett, J. A. (2020). Structure and functional analysis of the Legionella pneumophila chitinase ChiA reveals a novel mechanism of metal-dependent mucin degradation. PLoS Pathog 16(5): e1008342.
- Rondelet, A. and Condemine, G. (2013). Type II secretion: the substrates that won't go away. Res Microbiol 164(6): 556-561.
- Sturgill-Koszycki, S. and Swanson, M. S. (2000). Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J Exp Med 192(9): 1261-1272.
- Valdizan, M. C., Julian, J. and Carson, D. D. (1992). WGA-binding, mucin glycoproteins protect the apical cell surface of mouse uterine epithelial cells. J Cell Physiol 151(3): 451-465.
- White, R. C. and Cianciotto, N. P. (2019). Assessing the impact, genomics and evolution of type II secretion across a large, medically important genus: the Legionella type II secretion paradigm. Microb Genom 5(6): e000273.
- White, R. C., and Cianciotto, N. P. (2016). Type II secretion is necessary for optimal association of the Legionella-containing vacuole with macrophage Rab1B but enhances intracellular replication mainly by Rab1B-independent mechanisms.Infect Immun 84 (12): 3313-3327.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Grigoryeva, L. S., Rehman, S., White, R. C., Garnett, J. A. and Cianciotto, N. P. (2021). Assay for Assessing Mucin Binding to Bacteria and Bacterial Proteins. Bio-protocol 11(5): e3933. DOI: 10.21769/BioProtoc.3933.
Category
Microbiology > Microbe-host interactions > Bacterium
Microbiology > Microbial biochemistry > Protein > Interaction
Cell Biology > Cell-based analysis > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









