- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A High-throughput Interbacterial Competition Platform
Published: Vol 10, Iss 17, Sep 5, 2020 DOI: 10.21769/BioProtoc.3736 Views: 5823
Reviewed by: Gisela Di VenanzioGunjan MehtaXiaofeng Zhou

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Bacterial Competition Assay Based on Extracellular D-amino Acid Production
Laura Alvarez and Felipe Cava
Apr 5, 2018 9115 Views
Growth Recovery Assay and FACS-based Population Sorting Following Territorial Exclusion in Proteus mirabilis
Murray J. Tipping and Karine A. Gibbs
Mar 5, 2020 4996 Views
Abstract
Contact-dependent interbacterial competition is a common strategy used by bacteria to fight for their ecological niches. Interbacterial competition is monitored by a competition assay involving co-culturing the attacker and the recipient bacterial cells on agar, followed by recovery of the surviving recipient cells. Conventional interbacterial competition assays rely on serial dilution, plate spreading, and colony counting experiments for the readout. The high demand for time and labor in a competition assay limits its use for large-scale screening. However, a high-throughput interbacterial competition screening method is required to screen genetic factors involved in an interbacterial competition. Here, using Agrobacterium tumefaciens as an attacker and Escherichia coli as a recipient, we developed a robust, fast, efficient, and high-throughput type VI secretion system-dependent interbacterial competition screening platform. This system allows for 96 simultaneous competition assays without the need for serial dilution and plate spreading. Data analysis of this system relies on only direct and straightforward colony counting. This platform may be easily adapted to identify novel factors involved in any contact-dependent interbacterial competition systems.
Keywords: Competition assayBackground
Bacteria have evolved multiple strategies to fight other bacteria to gain ecological niche fitness, and contact-dependent interbacterial competition is one of the widely used strategies (Granato et al., 2019). A bacterium can use its type I (T1SS), type IV (T4SS), type V (T5SS), type VI (T6SS), or type VII (T7SS) secretion systems to deliver protein toxins into its competitor cell, the recipient cell, and kill it in a contact-dependent manner (Aoki et al., 2005; Hood et al., 2010; Souza et al., 2015; Cao et al., 2016; García-Bayona et al., 2017). The protein toxin-delivering bacterium is the attacker cell. The attacker cell also expresses the cognate immunity protein to protect itself from self-intoxication. A competition assay can measure the strength of a contact-dependent interbacterial competition. A competition assay can be divided into four steps: (1) culturing the attacker and recipient cells, (2) preparing the competition spot by mixing the two bacterial strains and incubating it on an agar plate, (3) recovery and selective growth of the recipient cells from the competition spot, and (4) counting the colony-forming units (CFUs) of the surviving recipient cells. The attacker with a robust interbacterial competition activity results in low recipient CFU.
Although the competition assay is a straightforward experiment, a high-throughput platform for 96 parallel competition assays is lacking. Several factors limit the capacity of a competition assay. First, each competition spot has to be separated at a distance to avoid cross-contamination during the recovery step. The need for the separation hinders the competition spots from arranging compactly, as in a 96-well format. Second, the recovery step and also the CFU counting step are time-consuming and laborious. In the recovery step, each competition spot is collected by an inoculating loop. Recording the CFU of the recipient cell requires serial dilution and plate spreading.
Here we present a high-throughput interbacterial competition platform that performs 96 competition assays simultaneously and does not require serial dilution and plate spreading. In this platform, the 96 competition mixtures were spotted on an agar plate solidified on a 96-well lid and recovered by a 96-pin microplate replicator instead of an inoculating loop. Using a 96-pin microplate replicator ensures that equal amounts of the bacteria are recovered, which is equivalent to the area of each pin. Because this method minimizes the amount of recovery to the pin area, serial dilution and plate spreading are not required. Meanwhile, the readout of this platform is direct and straightforward by counting the CFU of the recipient cells. This platform has been used to identify the recipient factors that are involved in enhancing the T6SS activity of Agrobacterium tumefaciens with success (Lin et al., 2020). We tested all 3,909 E. coli Keio mutants for their recovery after co-incubation with a T6SS active A. tumefaciens strain within 1 month. This method can be used to screen for both the attacker and recipient mutants with enhanced or reduced interbacterial competition activity mediated by any secretion system.
Materials and Reagents
- 250 μl tip for EzMate (KLBiotech, catalog number: FX-250-R )
- 96-pin microplate replicator (Violet BioScience, catalog number: VIO-T96 )
- Pipet tip box (Labcon, catalog number: 1055-965-018 )
- 96-well microplate, flat bottom (Basic Life, catalog number: BL6052 )
- 96-well microplate, U bottom (Basic Life, catalog number: BL6031 )
- 96-well lid (Basic Life, catalog number: BL6171 )
- 2.2-ml 96 deep-well microplate (Basic Life, catalog number: BL6038 )
- Test tube, 16 x 125 mm (Pyrex, catalog number: 9820 )
- Test tube cap (Kimble, catalog number: KIM-KAP 73660 )
- Pipetting reservoir (Basic Life, catalog number: BL6233 )
- A. tumefaciens C58 wild type (WT)
- Sucrose (Sigma-Aldrich, catalog number: S5016 )
- NZ-Case® Plus, casein enzymatic hydrolysate (Sigma-Aldrich, catalog number: N4642 )
- BactoTM Yeast extract (BD, catalog number: 212750 )
- K2HPO4 (Merck Millipore, catalog number: 7758-11-4 )
- MgSO4·7H2O (Merck Millipore, catalog number: 10034-99-8 )
- LB broth, Miller (BD, catalog number: 244620 )
- NaH2PO4·2H2O (Merck Millipore, catalog number: 10028-24-7 )
- NH4Cl (Merck Millipore, catalog number: 017-014-00-8 )
- KCl (Sigma-Aldrich, catalog number: P9541 )
- MES (Bio Basic, catalog number: 145224-94-8 )
- BactoTM Agar (BD, catalog number: 214010 )
- 95% ethanol (TaiSugar, catalog number: ethanol-18L )
- 6% bleach (LCY Chemical, catalog number: bleach-4L )
- Kanamycin sulfate (BioBasic, catalog number: KB0285-5g )
- HCl (Honeywell, catalog number: 30721-2.5L-GL )
- 523 broth (see Recipes)
- LB broth (see Recipes)
- LB agar with 20 μg/ml kanamycin (see Recipes)
- AK broth (see Recipes)
- AK agar, fresh prepare (see Recipes)
- 70% ethanol (see Recipes)
- 0.6% bleach (see Recipes)
Equipment
- Automated Pipetting System (KLBiotech, model: EzMateTM 401)
- Pipettes (Gilson, models: PIPETMAN P2, P20, P200, P1000, 8𝗑20)
- -80 °C freezer
- Centrifuge (Eppendorf, model: 5810R )
- Centrifuge rotor (Eppendorf, model: A-4-62 )
- Orbital shaking incubator (TKS, model: OSI-500R )
- Autoclave (LUXLEY, model: HL-326 )
- Microwave (Panasonic, model: NN-ST651 )
- 500 ml PYREX Erlenmeyer flasks (Corning, catalog number: 4980-500 )
- Alcohol burner (DG Life, catalog number: D92J-119250 )
Software
- EzStarter v4.0.0.87 (Arise Biotech)
Procedure
- Day 1: Pre-culture the attacker cells and the recipient cells
- Pre-culture the attacker cells.
Choose a single colony of A. tumefaciens C58 wild type (WT) grown on 523 agar and grow it in a test tube with 5 ml of 523 medium at 25 °C, 220 rpm for overnight. - Pre-culture the control recipient cells.
Choose a single colony of E. coli BW25113 WT carrying pRL-nptII plasmid and grow it in a test tube with 5 ml of LB medium with 20 μg/ml kanamycin. Grow the cells at 37 °C, 220 rpm overnight.
Note: The pRL-nptII provides BW25113 WT with kanamycin resistance so that it can be selectively grown after co-incubating with A. tumefaciens. - Sterilize the 96-pin microplate replicator.
- Place an alcohol burner on the left. Take a 200-μl tip box, remove the tip holder inside, flatten the box, and then put it on the right of the burner. Fill the left well of the flattened box with 0.6% bleach and the right well with sterilized water (Figure 1).
- Fill the 96-pin microplate replicator holder with 70% ethanol and place it on the right of the sterilized water-containing well (Figure 1).

Figure 1. Setup for sterilizing the microplate replicator. From left to right: alcohol burner, 0.6% bleach, sterilized water, and 70% ethanol. Place the alcohol burner distal to the 70% ethanol to avoid accidents.- Immerse the pins of the 96-pin microplate replicator serially in 0.6% bleach, sterilized water, and 70% ethanol. Let the replicator stand for 10 s in each solution.
- Sterilize the replicator with an alcohol burner (Figure 2) and cool the replicator for 10 s to room temperature. The sterilized replicator is ready to subculture E. coli mutants.
Note: Be very careful when handling the alcohol burner.

Figure 2. Sterilize the microplate replicator by flame. Place the 96-pin microplate replicator after its pins are immersed in 70% ethanol on top of the alcohol burner for sterilization. Move the replicator around the flame to make sure the alcohol on each pin is burned out, then cool the replicator for 10 s at room temperature. The sterilized replicator is ready to subculture E. coli mutants. - Pre-culture the E. coli mutants from the Keio library that serve as recipient cells.
- Take a U-bottom 96-well microplate and fill each well with 100 μl LB medium with 20 μg/ml kanamycin.
- Take out an E. coli Keio mutant plate, which is stored as glycerol stock in a 2.2-ml 96 deep-well format in a -80 °C refrigerator. Apply the sterilized replicator to the frozen E. coli stocks.
Note: Make sure each replicator pin contacts the surface of the glycerol stock. - Stamp the bacteria-containing replicator onto the LB medium in each well of the 96-well microplate prepared in Step A4a and agitate the liquid to resuspend the bacteria thoroughly (Figure 3).
Note: Make sure each replicator pin contacts the LB medium in each well of a 96-well plate.
Figure 3. Pre-culture the E. coli mutants from the Keio library. Stamp the bacteria-containing replicator onto LB medium in each well of a 96-well microplate and agitate the liquid to resuspend the bacteria thoroughly. - Reseal and return the E. coli Keio mutants plate back to -80 °C and place the replicator in the 0.6% bleach prepared in Step A3a.
- Add a 96-well lid to the E. coli-containing U-bottom 96-well microplate and incubate at 37 °C, 220 rpm for 16 h (Figure 4).
Note: The incubation can be achieved by adding the anti-slip pad on a regular incubator.
Figure 4. Incubating the E. coli-containing U-bottom 96-well microplate in an incubator. Remove some of the flask holders and replace the space with an anti-slip pad. Fix the anti-slip pad with screws. The anti-slip–containing area is ready for incubating the 96-well microplate.
- Sterilize the 96-pin microplate replicator (Steps A3c to A3d).
- Pre-culture the attacker cells.
- Day 2: Sub-culture the attacker cells and the recipient cells
Note: This step incorporates BW25113(pRL-nptII) into the Keio mutant-containing microplate and synchronizes their growth.- Sub-culture the attacker cells.
Sub-culture 1 ml of the A. tumefaciens C58 WT from Step A1a with 100 ml of 523 medium in a 500-ml flask, prepare two cultures (200 ml in total). Grow the bacteria at 25 °C, 220 rpm, overnight. - Sub-culture the recipient cells.
- Take a U-bottom 96-well microplate and fill each well with 100 μl LB medium with 20 μg/ml kanamycin. Use an 8-channel pipette to subculture 2 μl E. coli mutants grown from Step A4e in the LB-containing U-bottom 96-well microplate but leave some wells empty for the control strain, BW25113(pRL-nptII) (Figure 5).
Note: Remove appropriate tips from the 8-channel pipette while sub-culturing to leave the well empty for the control strain. - Sub-culture 2 μl BW25113(pRL-nptII) from Step A2 into the empty wells in Step B2a (WT label in Figure 5).
- Grow the sub-cultured E. coli strains at 37 °C, 220 rpm, for overnight.
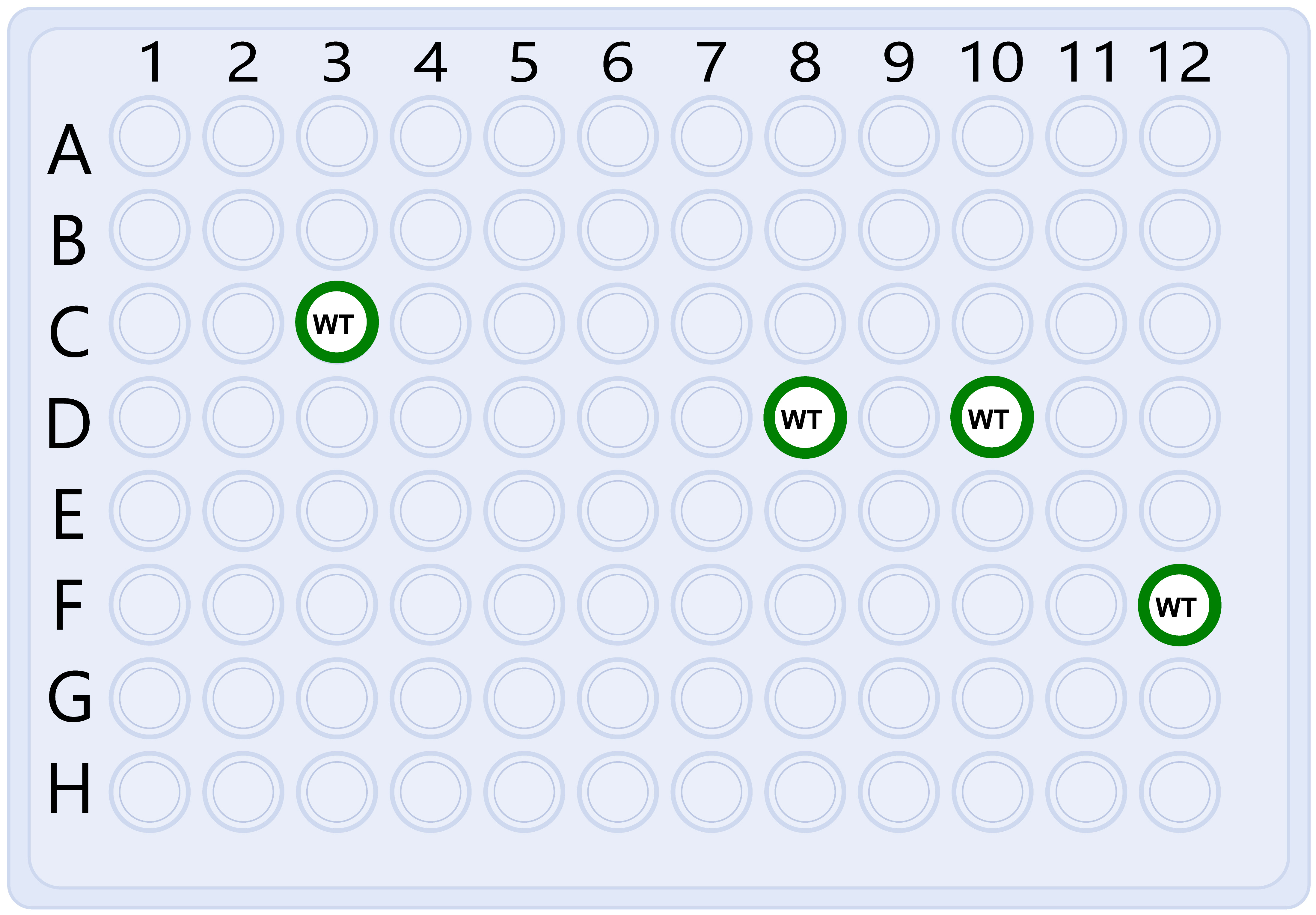
Figure 5. Sub-culture the E. coli Keio mutants and the BW25113(pRL-nptII). Use an 8-channel pipette to subculture 2 μl of the overnight-cultured E. coli mutants from step A4e into a new plate filled with 100 μl LB medium with 20 μg/ml kanamycin. Do not sub-culture the wells reserved for BW25113(pRL-nptII) controls (e.g., Wells 3C, D8, D10, and F12 in this case). Sub-culture 2 μl overnight-cultured BW25113(pRL-nptII) into the empty wells as controls (e.g., wells labeled WT).
- Take a U-bottom 96-well microplate and fill each well with 100 μl LB medium with 20 μg/ml kanamycin. Use an 8-channel pipette to subculture 2 μl E. coli mutants grown from Step A4e in the LB-containing U-bottom 96-well microplate but leave some wells empty for the control strain, BW25113(pRL-nptII) (Figure 5).
- Sub-culture the attacker cells.
- Day 3: Co-incubate the attacker cells and the recipient cells to enable interbacterial competition
- Prepare the competition surface.
Pour 25 ml melted AK agar on a 96-well lid in the laminar flow and allow it to dry for 45-60 min.
Note: It is crucial to fix the drying time because surface dryness affects competition outcomes. - Adjust the OD600 of the attacker cells.
- Centrifuge the 200 ml cultured A. tumefaciens C58 WT at 8,000 x g for 10 min. Discard the supernatant and resuspend the cell pellet with 20 ml of 0.9% NaCl.
- Centrifuge the A. tumefaciens cells at 5,000 x g for 5 min. Discard the supernatant and resuspend the cell pellet with 10 ml of 0.9% NaCl.
- Measure the OD600 of the washed A. tumefaciens cells and adjust to OD600 = 3.0 with 0.9% NaCl.
- Aliquot attacker cells to a 2.2 ml 96 deep-well microplate using the automated pipetting system.
- Open the automated pipetting system and the EzStarter software that controls the operation of the EzMate automated pipetting system. Set the software to aliquot 150 μl of the sample in Block R1 into each well of Block A; repeat the procedure once so that each well contains 300 μl liquid (Figure 6A)
- Dispense 50 ml of the OD600-adjusted A. tumefaciens to a pipetting reservoir and place the reservoir in Block R1, a sterilized 2.2 ml 96 deep-well microplate in block A, and a box of EzMate 250 μl tip in block D (Figure 6B).
- Use the automated pipetting system to aliquot 150 μl A. tumefaciens into each well of the 2.2-ml 96 deep-well microplate. Repeat the cycle once so that each well contains 300 μl A. tumefaciens (Figure 6C).

Figure 6. Aliquot attacker cells (A. tumefaciens) to a 2.2-ml 96 deep-well microplate. A. Set the EzStarter program for dispensing 150 μl of the liquid in Block R1 in each well of Block A twice. B. Set up the working station by placing the A. tumefaciens-containing pipetting reservoir in Block R1, a 2.2 ml 96 deep-well microplate in Block A, and a box of EzMate 250 μl tip in Block D. C. The automated pipetting system dispenses A. tumefaciens to Block A.
- Mix the attacker and recipient cells and subject the mixture to AK agar to start the interbacterial competition.
- Set up the automated pipetting system.
Remove the A. tumefaciens-containing pipetting reservoir from Block R1 and let the 2.2-ml 96 deep-well microplate with 300 μl A. tumefaciens in each well (from Step C3c) remain in Block A. Take out the overnight-subcultured E. coli plate (from Step B2c) and place it in Block C. Place the competition surface (from Step C1) in Block B. Place a box full of 250 μl tip for EzMate in Block D (Figure 7A). - Use the automated pipetting system to add 10 μl overnight-cultured E. coli (Block C) into the A. tumefaciens-containing 2.2-ml 96 deep-well microplate (Block A). Mix the bacterial suspension 10 times, and drop 10 μl mixture onto the AK plate (Block B) (Video 1).Video 1. Mix the attacker and recipient cells and subject the mixture to AK agar. Use the automated pipetting system to add 10 μl overnight-cultured (upper right Block C) into the A. tumefaciens-containing 2.2-ml 96 deep-well microplate (upper left Block A). Mix the bacterial suspension 10 times and drop 10 μl mixture onto the AK plate (lower left Block B).
- Repeat each column until all 96 wells are mixed and dispensed (Figure 7B).
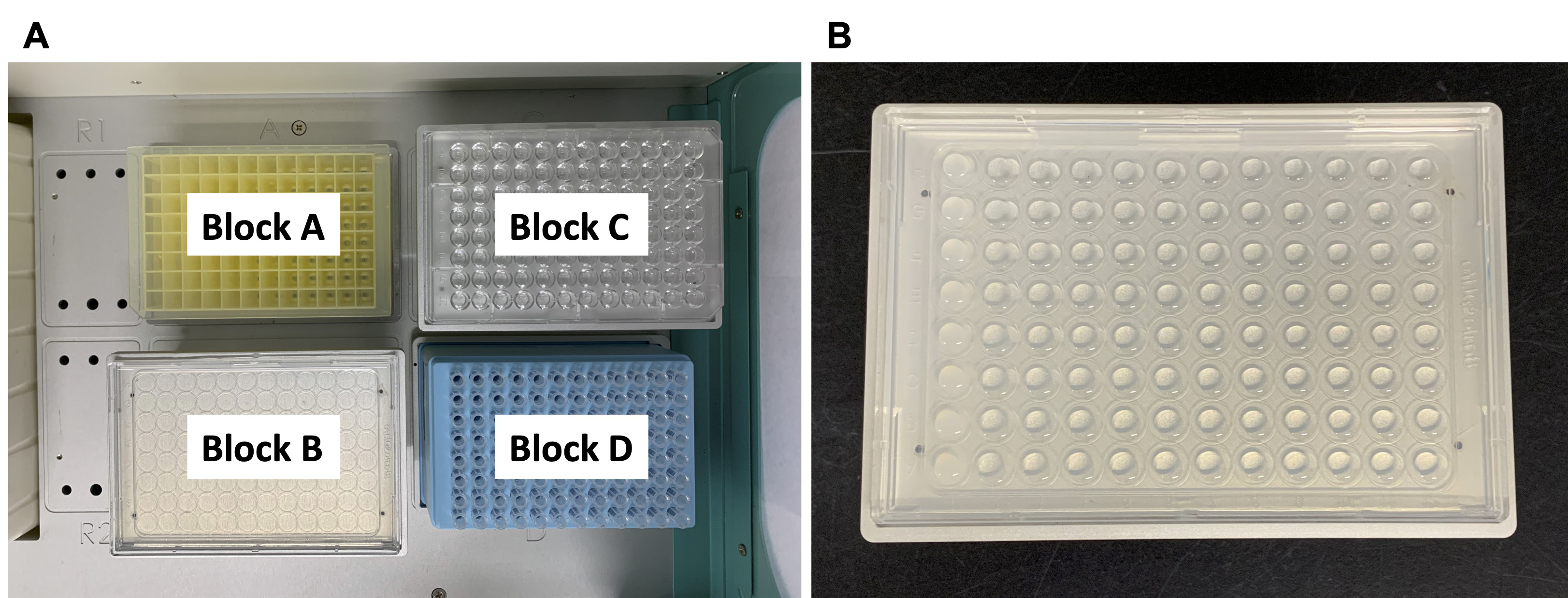
Figure 7. Mix the attacker and recipient cells and subject the mixture to the competition surface (AK agar). A. Set up of the working station. Block A contains the 2.2-ml 96 deep-well microplate with 300 μl A. tumefaciens in each well (from Step C3c), Block C on the upper right contains the overnight-subcultured E. coli (from Step B2C), Block B on the lower left contains the competition surface (from Step C1), and Block D contains a full box of 250 μl tip for EzMate. B. Competition surface after dispensing 10 μl attacker/recipient mixture. Make sure all liquid mixture has dried before incubating it at 25 °C for 16 h. - Dry the bacteria-containing AK agar in a laminar flow until all the competition spots are dry (about 15 min). Cover the dried AK agar with another 96-well lid. Place the plate at 25 °C for 16 h to allow competition to take place (Figure 8).

Figure 8. The competition surface after 16 h of incubation - Set up the automated pipetting system.
- Prepare the competition surface.
- Day 4: Recover the competition spots
- Prepare the recovery plate.
Pour 25 ml melted LB agar supplied with 20 μg/ml kanamycin on a 96-well lid in the laminar flow and allow it to dry for 45-60 min.
Note: This procedure is the same for Step C1 but uses a different medium. - Prepare the recovering liquid.
Fill each well of a U-bottom 96-well plate with 200 μl of 0.9% NaCl. - Sterilize the 96-pin microplate replicator as in Step A3.
- Recover bacterial cells from the competition spot.
- Place the sterilized replicator onto the competition surface (Figure 9A).
Note: Make sure each pin contacts one competition spot. - Lift the replicator (Figure 9B) and immerse the pins in the recovery liquid prepared in Step D2. Swirl and agitate the liquid to thoroughly resuspend the bacterial cells (Figure 9C).
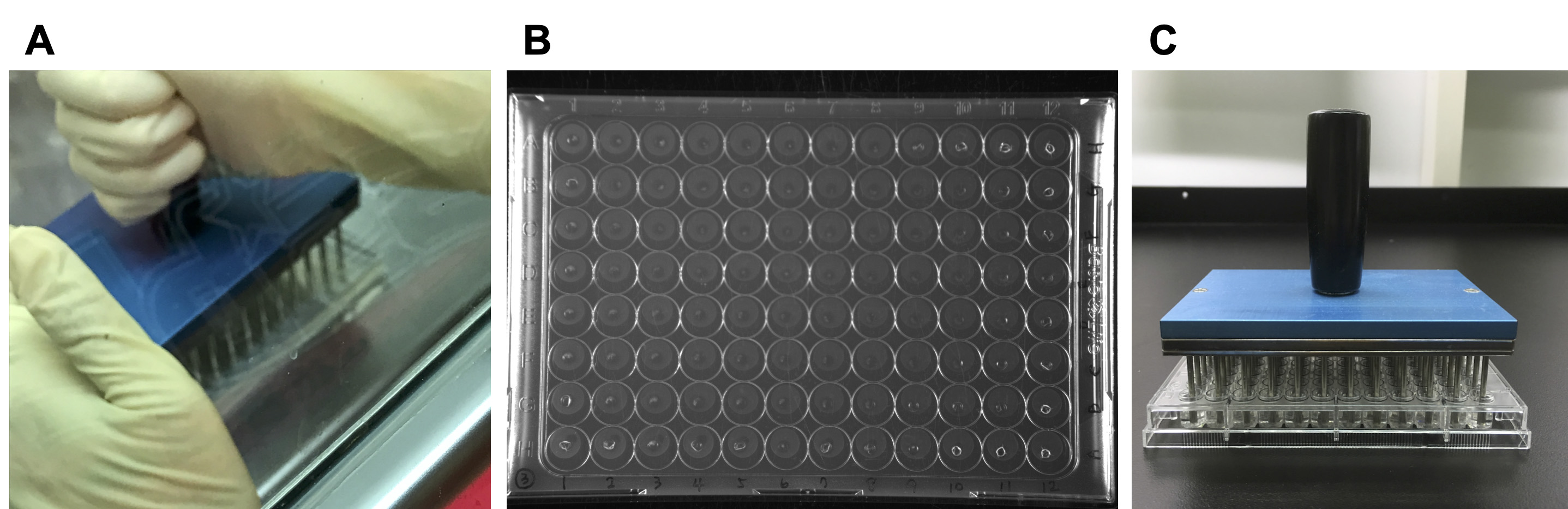
Figure 9. Recover the interbacterial competing bacterial cells from the AK agar. A. Stamp the bacterial mixture by a sterilized microplate replicator. B. The bacteria-containing AK agar should have an apparent hole in each spot after stamping. C. Resuspend the bacterial mixture into 200 μl of 0.9% NaCl.
- Place the sterilized replicator onto the competition surface (Figure 9A).
- Spot the recovered bacterial mixture onto the LB agar with 20 μg/ml kanamycin.
- Set up the automated pipetting system demonstrated in Figure 10.
Place the bacteria-containing U-shape 96-well plate (from Step D4b) in Block A, the recovery plate (from Step D1) in Block B, and a box filled with 250 μl tip for EzMate in Block D (Figure 10).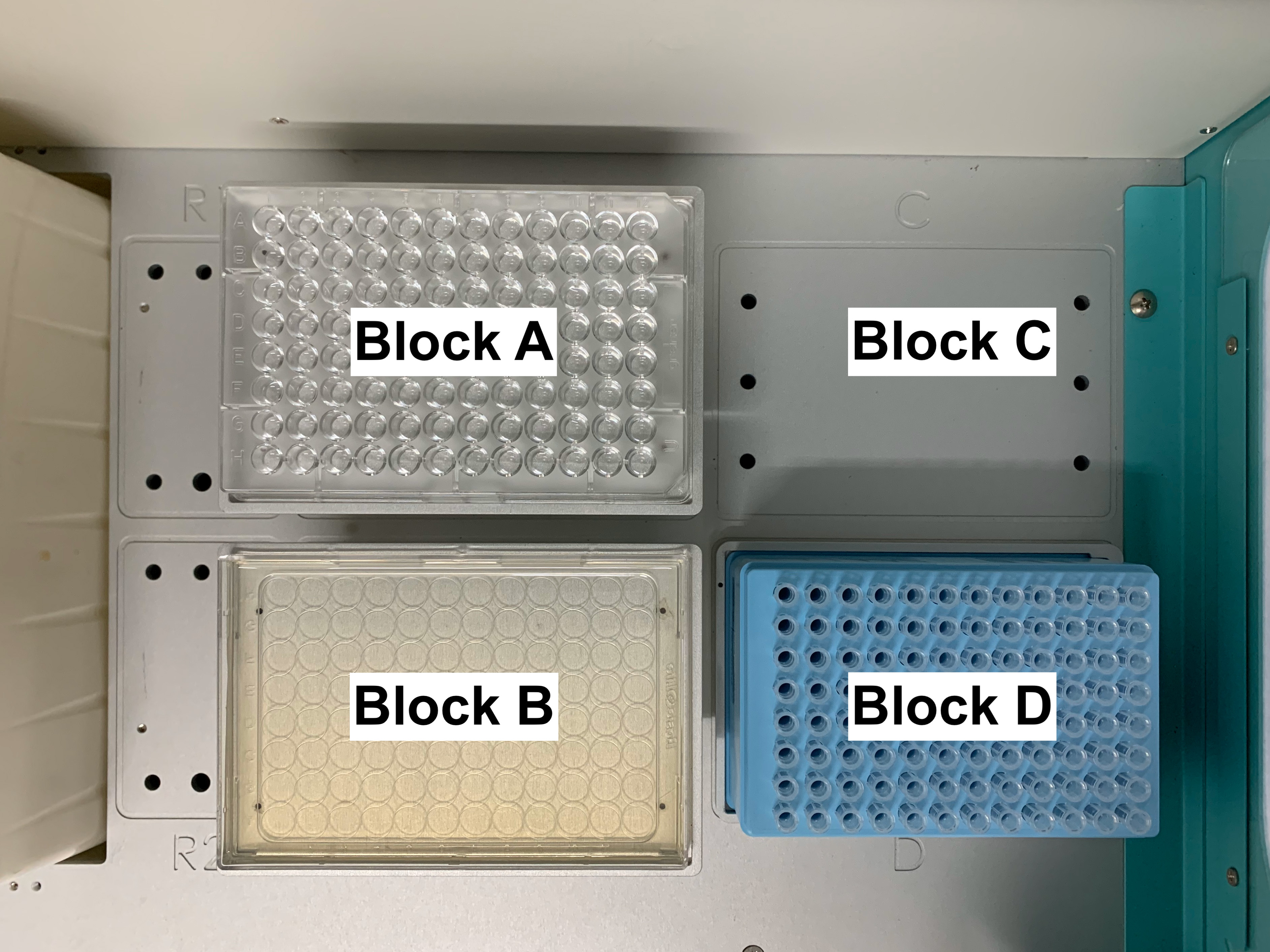
Figure 10. Set up for spotting the recovered bacteria onto LB agar with 20 μg/ml kanamycin. Place the bacteria-containing U-shape 96-well plate (from Step D4b) in Block A, the recovery plate (from Step D1) in Block B, and a box filled with 250 μl tip for EzMate in Block D. - Use the automated pipetting system to mix the recovered bacterial mixture 10 times (Block A) and transfer 10 μl of the recovered bacterial mixture (Block A) to the recovery plate (Block B) (Video 2).Video 2. Transfer 10 μl of the recovered bacteria to the recovery plate. Mix the recovered bacterial suspension 10 times (Block A) and transfer 10 μl of the recovered bacterial mixture (Block A) to the recovery plate (Block B). Repeat each column until all 96 wells are mixed and dispensed.
- Repeat each column until all the 96 wells are mixed and dispensed.
- Set up the automated pipetting system demonstrated in Figure 10.
- Selectively grow the recipient cells.
Dry the plate in a laminar flow until all spots are dry (takes about 15 min). Cover the dried plate with another 96-well lid. Place the plate at 37 °C overnight to allow recipient cells to grow.
Note: All liquid must be dried out before being placed in an incubator. The drying time of each plate should be the same.
- Prepare the recovery plate.
- Day 5: Counting the colony-forming unit (CFU) of the surviving recipient cells.
Take a picture of the plate for data preservation. The wells with BW25113(pRL-nptII) as the recipient cell should yield no or only 1-2 colonies. Thus, the wells with multiple colonies are the E. coli mutant candidates that are less susceptible to A. tumefaciens T6SS attack.
Data analysis
- Check the survival rate of E. coli BW25113(pRL-nptII) control strain in each well (green circle, Figure 11), and all control wells should contain no more than 2 colonies.
- Record the data by taking a photo of the recovery plate.
- Observe the number of surviving colonies directly without the need for any equipment or software. The wells with multiple colonies (orange circle, Figure 11) are the E. coli Keio mutants that are less susceptible to A. tumefaciens antibacterial killing.
Note: The wells with more than 7 colonies are considered less susceptible candidates and are selected as resistant candidates for further validation.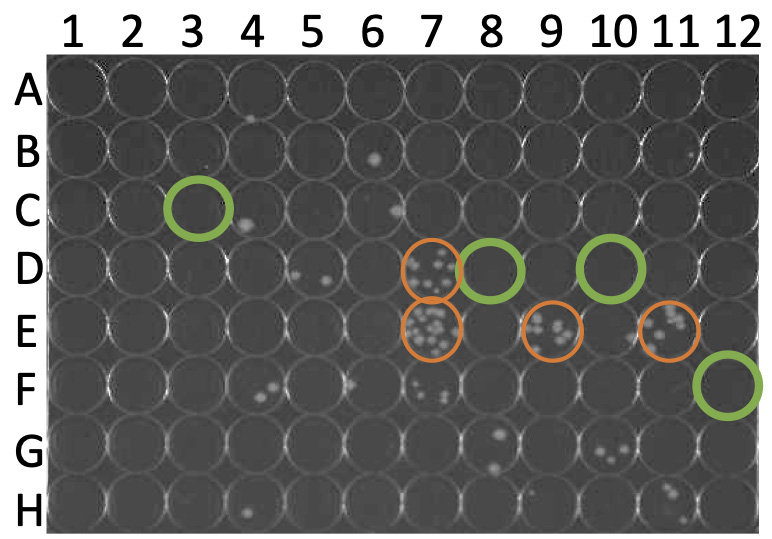
Figure 11. Recovery plate readout. The wells with green circles carry control recipient cells (E. coli BW25113 WT carrying pRL-nptII). The wells with orange circles are the candidate strains that are less susceptible to A. tumefaciens antibacterial killing.
Notes
- This protocol describes a high-throughput screening system to identify genetic factors of recipient cells affecting the T6SS-mediated interbacterial competition outcome. However, this method can be adapted to any contact-dependent interbacterial competition system.
- The automated pipetting system used in this protocol is EzMate401, but any automated pipetting system can be used and even can be adapted by an 8-channel pipette.
Recipes
- 523 broth
- Dissolve 10 g sucrose, 8 g casein enzymatic hydrolysate, 4 g yeast extract, 3 g K2HPO4, and 0.3 g MgSO4·6H2O in 800 ml distilled water
- Adjust pH to 7.0 with HCl then bring the volume to 1,000 ml with distilled water
- Autoclave the solution at 121 °C for 20 min
- Store the autoclaved 523 broth at room temperature
- LB broth
- Dissolve 25 g LB powder in 1,000 ml distilled water
- Autoclave the solution at 121 °C for 20 min
- Store the autoclaved LB broth at room temperature
- LB agar with 20 μg/ml kanamycin
- Dissolve 1.5 g agar in 100 ml LB broth
- Autoclave the solution at 121 °C for 20 min
- Cool the autoclaved media and add kanamycin to a final concentration of 20 μg/ml
- Take 25 ml of the media using a 25 ml pipette and pour 20 ml into a 96-well cover
- Spread the agar evenly and wait until the agar solidifies
- Store the autoclaved LB agar at 4 °C
- AK broth
- Dissolve 3 g K2HPO4, 1 g NaH2PO4, 1 g NH4Cl, 0.15 g KCl, and 9.76 g MES in 900 ml distilled water
- Adjust pH to 5.5 with NaOH then bring the volume to 1,000 ml with distilled water
- Autoclave the solution at 121 °C for 20 min
- Store the autoclaved AK broth at room temperature
- AK agar, fresh prepared
- Dissolve 2 g agar in 100 ml AK broth
- Melt the agar by microwave to heat the solution until all the agar dissolves
- Mix the media well and cool at room temperature for 2 min
- Take 25 ml hot media by using a 25-ml pipet and pour 20 ml into a 96-well cover
- Spread the agar evenly and wait for 1 h for the agar to solidify and dry the surface
- 70% ethanol
- Mix 737 ml of 95% ethanol with 263 ml sterile water
- Store at room temperature
- 0.6% bleach
- Mix 10 ml of 6% bleach with 100 ml sterile water
- Store at room temperature, avoid light contact
Acknowledgments
The authors thank Jemal Ali, Hagos Mohammedseid Juhar, and Maria Karmella Apaya for critically reading this protocol. This work reports a detailed protocol used to identify the E. coli recipient factors that are involved in enhancing the T6SS activity of A. tumefaciens C58 (Lin et al., 2020). The funding for this project was provided by the Ministry of Science and Technology of Taiwan (MOST) (grant no. 104-2311-B-001-025-MY3) and Academia Sinica Investigator Award (grant no. AS-IA-107-L01) to E-ML. The E. coli Keio collection library was provided by the National BioResource Project (NIG, Japan): E. coli.
Competing interests
The authors declare no conflicts of interest.
References
- Aoki, S. K., Pamma, R., Hernday, A. D., Bickham, J. E., Braaten, B. A. and Low, D. A. (2005). Contact-dependent inhibition of growth in Escherichia coli. Science 309(5738): 1245-1248.
- Cao, Z., Casabona, M. G., Kneuper, H., Chalmers, J. D. and Palmer, T. (2016). The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat Microbiol 2(1): 16183.
- García-Bayona, L., Guo, M. S. and Laub, M. T. (2017). Contact-dependent killing by Caulobacter crescentus via cell surface-associated, glycine zipper proteins. eLife 6: 24869.
- Granato, E. T., Meiller-Legrand, T. A. and Foster, K. R. (2019). The evolution and ecology of bacterial warfare. Curr Biol 29(11): R521-R537.
- Hood, R. D., Singh, P., Hsu, F., Güvener, T., Carl, M. A., Trinidad, R. R., Silverman, J. M., Ohlson, B. B., Hicks, K. G., Plemel, R. L., Li, M., Schwarz, S., Wang, W. Y., Merz, A. J., Goodlett, D. R. and Mougous, J. D. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host & Microbe 7(1): 25-37.
- Lin, H.-H., Yu, M., Sriramoju, M. K., Hsu, S.-T. D., Liu, C.-T. and Lai, E.-M. (2020). A High-throughput interbacterial competition screen identifies ClpAP in enhancing recipient susceptibility to type VI secretion system-mediated attack by Agrobacterium tumefaciens. Front Microbiol 10(3077).
- Souza, D. P., Oka, G. U., Alvarez-Martinez, C. E., Bisson-Filho, A. W., Dunger, G., Hobeika, L., Cavalcante, N. S., Alegria, M. C., Barbosa, L. R. S., Salinas, R. K., Guzzo, C. R. and Farah, C. S. (2015). Bacterial killing via a type IV secretion system. Nat Commun 6(1): 6453.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lin, H. and Lai, E. (2020). A High-throughput Interbacterial Competition Platform. Bio-protocol 10(17): e3736. DOI: 10.21769/BioProtoc.3736.
Category
Microbiology > Microbial physiology > Interspecific competition
Microbiology > Microbial signaling > Interspecies communication
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












