- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
The ATPase Activity of Escherichia coli Expressed AAA+-ATPase Protein
Published: Vol 10, Iss 15, Aug 5, 2020 DOI: 10.21769/BioProtoc.3705 Views: 4902
Reviewed by: Zhibing LaiJoyce ChiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
Kevin Baudry and Emmanuelle Issakidis-Bourguet
Aug 20, 2023 1463 Views
An in vitro Assay to Probe the Formation of Biomolecular Condensates
Yu Zhang and Shen Lisha
Sep 5, 2023 3190 Views
Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1806 Views
Abstract
ATPases are the enzymes that breakdown ATP to ADP and release inorganic phosphate (Pi). Here we provide a detailed protocol to determine the ATPase activity of a recombinant AAA+-ATPase protein (GENERAL CONTROL NON-REPRESSIBLE-4 [GCN4]) by spectrophotometric absorption at 360 nm to measure the accumulated inorganic phosphate. In general, the substrate 2-amino-6-mercapto-7-methylpurine riboside (methylthioguanosine, a guanosine analog: MESG) is enzymatically converted in the presence of Pi by purine nucleoside phosphorylase (PNP) to ribose 1-phosphate and 2-amino-6-mercapto-7-methylpurine. The spectrophotometric shift in maximum absorbance at 330 nm for the MESG substrate and subsequent conversion product at 360 nm due to enzymatic conversion was measured. The GCN4-His-tagged recombinant protein was expressed in Escherichia coli BL21 cells and purified using Ni-NTA column. This purified protein was then used for the quantitation of Pi in solution or the continuous determination of Pi released due to the ATPase activity of GCN4, an AAA+-ATPase protein conserved in many eukaryotes, which in plants regulates stomatal aperture during biotic and abiotic stress in plants.
Background
Adenosine triphosphatases (ATPases) are a class of enzymes that catalyze the breakdown of ATP into ADP and free inorganic phosphate (Pi). This breakdown and release of Pi generates energy used by enzymes to carry out the chemical reactions that require energy. This process is an integral part of all the kingdom of life. ABC transporters are the transmembrane proteins that move solute through the membrane by ATPase activity (Rees et al., 2009). Some ATPases are cytoplasmic or membrane-associated proteins. AAA+ proteins are the ATPases associated with diverse cellular activities such as protein degradation, membrane fusion, disassembly of protein complexes, microtubule dynamics, etc. (Snider et al., 2008). We identified GENERAL CONTROL NON-REPRESSIBLE-4 (GCN4), an AAA+-ATPase, as a novel player in regulating stomatal aperture and thus playing a role in plant innate immunity and drought tolerance (Kaundal et al., 2017). Here we describe the method to prove its ATPase activity. The GCN4 as an ATPase enzyme converts ATP to ADP and Pi, and then the enzyme phosphoribosyl phosphorylase (PNP) converts the MESG substrate into the final substrate. The assay monitors the spectrophotometric shift of the substrate (MESG) in the presence of Pi from 330 to 360 nm. The conversion rate is directly correlated with the amount of Pi present in the solution and, that depends on the conversion of ATP to ADP by GCN4 ATPase activity. We reported the ATPase activity of a recombinant protein (GCN4-His) as absorbance (ABS) at 360 nm vs. protein concentration (Kaundal et al., 2017).
Many techniques have been used to quantify the in vitro ATPase activity of purified proteins. Most of the techniques for quantification are based on the detection of free inorganic phosphate (Pi). The most common method uses the calorimeter substrate malachite green (Carter and Karl, 1982). Besides this, fluorescent and radioactive substrates are also used in various protocols to detect free phosphate in the ATPase reaction (Brune et al., 1994; Shiue et al., 2006). In this protocol, we used 2-amino-6-mercapto-7-methylpurine riboside (methylthioguanosine, a guanosine analog: MESG) as a substrate to detect the presence of free phosphate during ATPase activity (Webb, 1992). The purine nucleoside phosphorylase (PNP) enzyme in the presence of inorganic phosphate, which is released from ATP (an experimental substrate) upon hydrolysis to ADP by ATPase (experimental enzyme to be tested), converts MSEG (assay substrate) to ribose 1-phosphate and 2-amino-6-mercapto-7-methylpurine. This enzymatic conversion of MSEG results in a spectrophotometric shift in maximum absorbance from 330 nm for the MESG substrate to 360 nm for the product 2-amino-6-mercapto-7-methylpurine (Figure 1). The advantage of this protocol is that it does not require long incubation steps and the reaction can be incubated in the plate reader itself for the required duration. The protocol is based on the ENZcheck Phosphate Assay Kit. Most of the components for enzyme assay come with the kit, including potassium phosphate standard, so the reaction is very convenient to assemble. We optimized the protocol from 1 ml reaction to 300 µl reaction to carry out in a microtiter plate. This protocol describes the detailed assay for calculating the ATPase activity of a recombinant GCN4, an AAA+-ATPase protein in U/ml, and specific activity. This protocol can also be used for the calculation of GTPase activity of a protein by using GTP as a substrate (Webb and Hunter, 1992).

Figure 1. Enzymatic conversion of 2-amino-6-mercapto-7-methylpurine riboside (methylthioguanosine, a guanosine analog: MESG) into ribose 1-phosphate and 2-amino-6-mercapto-7-methylpurine by purine nucleoside phosphorylase (PNP) in the presence of inorganic phosphate released from ATP (substrate) by ATPase (enzyme to be tested)
Materials and Reagents
- 96-well microtiter plate (BD Biosciences, catalog number: 353075 )
- EnzCheck Phosphate Assay Kit (Thermo Fisher, catalog number: E6646 )
- ATP (Sigma, catalog number: A2383 )
- ATPase (Sigma, catalog number: A7510 )
- Bio-Rad Protein Assay (Bio-Rad Laboratories, catalog number: 500-0006 )
- Bovine Serum Albumin (BSA) 50 mg/ml (InvitrogenTM, catalog number: 15561020 )
- Tris base
- Recombinant ATPase protein/enzyme you would like to test (we used GCN4-HisTag Fusion protein and referred to as test protein below)
- 1 M Tris-Cl (pH-8.0) (see Recipes)
Equipment
- Microtiter plate reader (Tecan, model: Infinite M200 Pro )
- Water bath (Thermo Scientific, catalog number: TSGP02 )
- Vortex (FisherBrands, catalog number: 14-955-163 )
- Autoclave
Procedure
- Estimation of protein concentration by Bradford assay (Bradford, 1976)
- Prepare a 100 µg/ml stock solution of BSA in water from BSA 50 mg/ml.
- Use a 96-well microtiter plate to prepare the reaction mix. Make triplicates for all reactions.
- Prepare 2.5, 5, 10, 20, and 50 µg of BSA standard in corresponding well as described in Table 1 below:
Table 1. Protein estimation reaction mix composition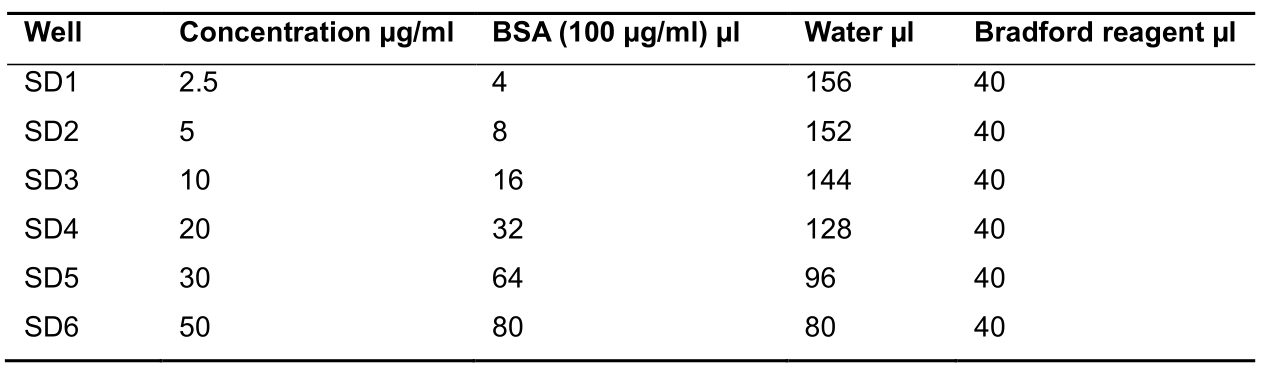
- Prepare a blank by adding 160 µl of water in a blank microtiter plate well.
- Prepare test samples in a microtiter plate well by making several dilutions of purified recombinant ATPase protein/enzyme with water to make a total of 160 µl volume.
- Add 40 µl of Bradford reagent to each microtiter plate well, mix and incubate for 5 min at room temperature.
- Read absorbance (ABS) at 595 nm on a microtiter plate reader.
- If the spectrophotometer does not have software to plot the standard curve, then plot the standard curve in Excel by plotting known BSA concentrations on X-axis and ABS595 on Y-axis. Obtain trendline and use the equation for the trendline and the ABS595 of the unknown to resolve the unknown concentration. The representative data are shown in Figure 2.
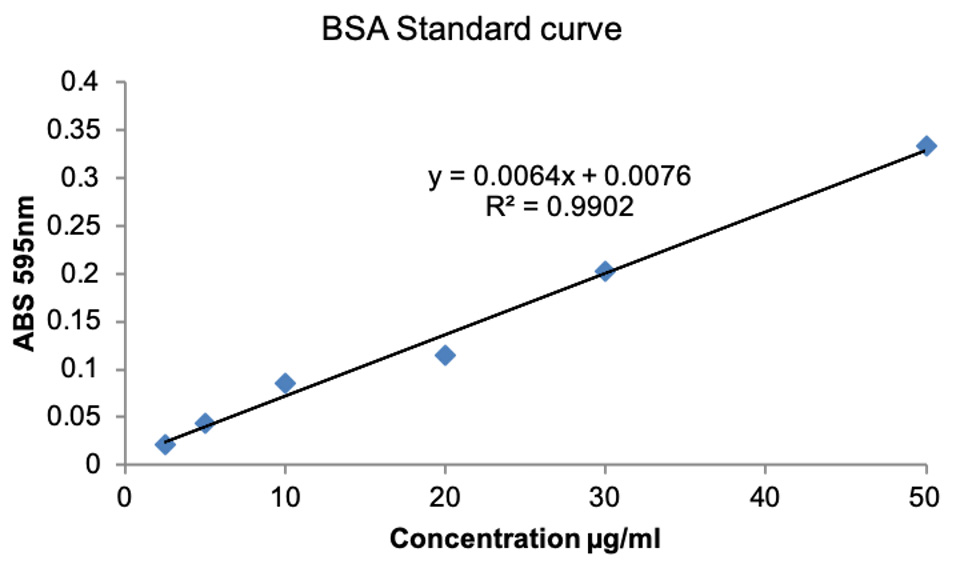
Figure 2. BSA standard curve
- ATPase assay
Reagent preparation
Prepare stock solutions supplied with the EnzCheck Phosphate Assay Kit to perform the assay.- Prepare a 1 mM stock solution of the MESG substrate by adding 20 ml of dH2O directly to the bottle (Component A). Mix extensively to dissolve MESG completely (maybe ~10 min).
Note: Do not heat to dissolve. Because MESG is near its saturation point, a small amount of solid may remain, even after extensive mixing. Immediately after dissolving the MESG substrate, aliquot the solution into convenient volumes and place immediately at -20 °C. - Thaw an aliquot of MESG substrate before use by placing it in a 37 °C water bath until just melted (not more than 5 min). Vortex vigorously and immediately chill the solution by placing it on ice.
Note: The solution is stable for at least 4 h on ice at pH 7.5. If left at room temperature, the half-life of MESG is about 4 h at pH 8.0 and 40 h at pH 6.0. Do not refreeze leftover MESG substrate. - Prepare enzyme purine nucleoside phosphorylase (Component B) stock by adding 500 µl of dH2O to a vial to prepare a 100 U/ml stock solution and store at 4 °C.
- Prepare recombinant ATPase protein (test protein) stock.
Note: Use different concentrations of test protein to analyze ATPase activity. - Prepare 1 mM ATP stock. Make 10 mM ATP stock by adding 0.05 g of ATP (disodium salt) in 10 ml of 25 mM Tris-Cl (pH 8.0) buffer (250 µl of 1 M Tris-Cl pH 8.0 in 9.75 ml sterilized water). Dilute to 1 mM ATP stock by adding 1 ml of 10 mM ATP in 9 ml of 25 mM Tris-Cl (pH 8.0).
- Dilute a portion of the 50 mM potassium phosphate monobasic (KH2PO4), a phosphate standard, 100-fold with dH2O to generate a 500 µM solution. Further, dilute a portion of 500 µM solution to make 1 µM KH2PO4, working solution.
Phosphate Standard curve- Prepare 10, 20, 30, 40, and 50 nmol of inorganic phosphate standard with 1 µM KH2PO4 in corresponding well to the final volume of 300 µl reaction mix as per Table 2 below:
Table 2. Phosphate standard curve reaction mix composition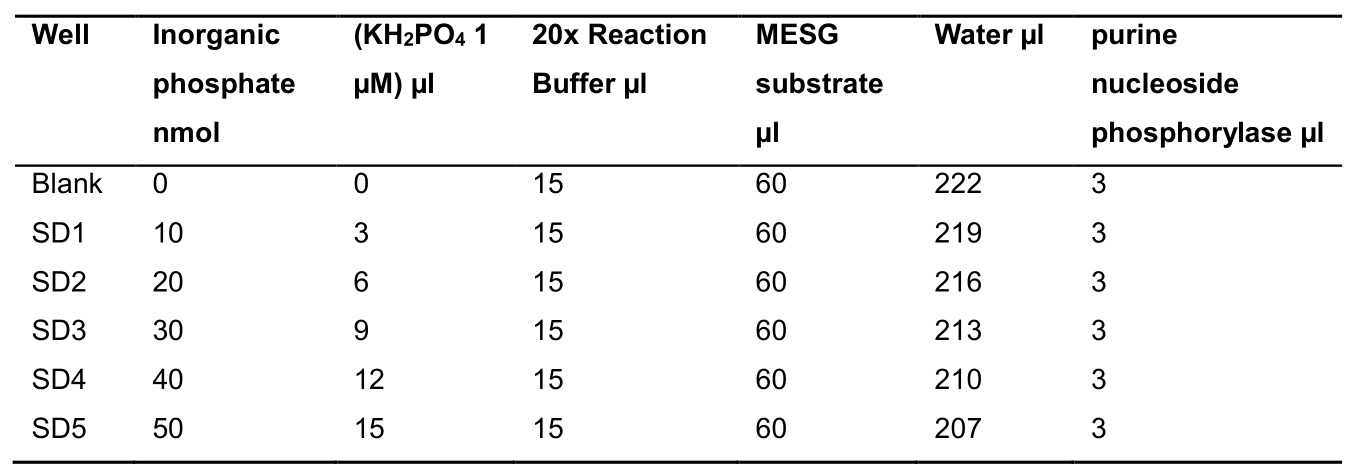
- Use a 96-well microtiter plate to perform the assay.
- Start the reaction by the addition of final component purine nucleoside phosphorylase and incubate for 30 min at 22 °C.
- Read the absorbance at 360 nm. Correct the absorbance by subtracting blank from each standard.
- Plot Phosphate standard curve in Excel by plotting known inorganic phosphate concertation on X-axis and ABS360 on Y-axis and obtain trendline and equation. The representative data is shown in Figure 3.
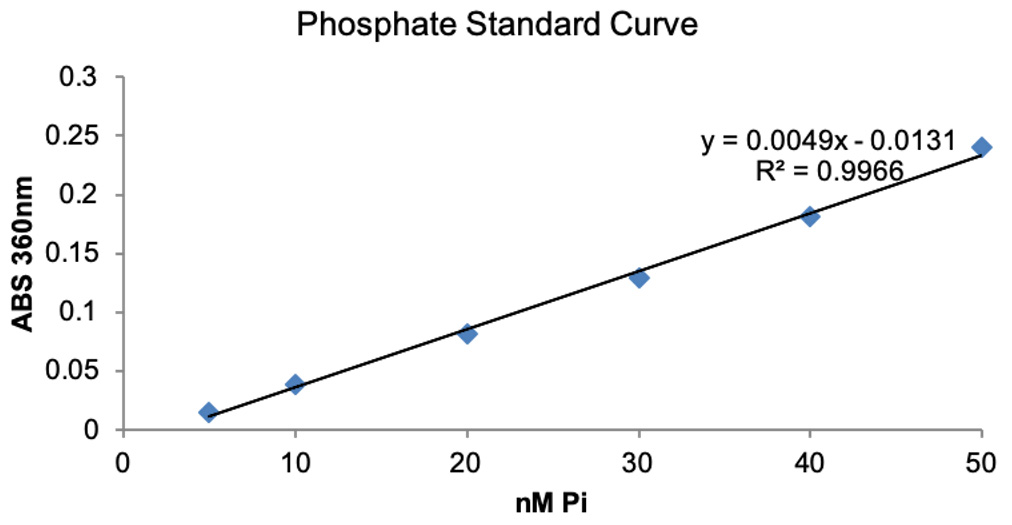
Figure 3. Phosphate standard curve
- Use a 96-well microtiter plate to perform the assay. If the plate reader is not available, use 1 ml of assay mix to perform the assay in cuvettes.
- Make triplicate for all reactions.
- Prepare assay mix by mixing the stock solution to make 300 µl assay mix for plate reader in 96-well plates and 1 ml assay mix to use in the cuvette as described in Table 3 below:
Table 3. ATPase assay reaction mix composition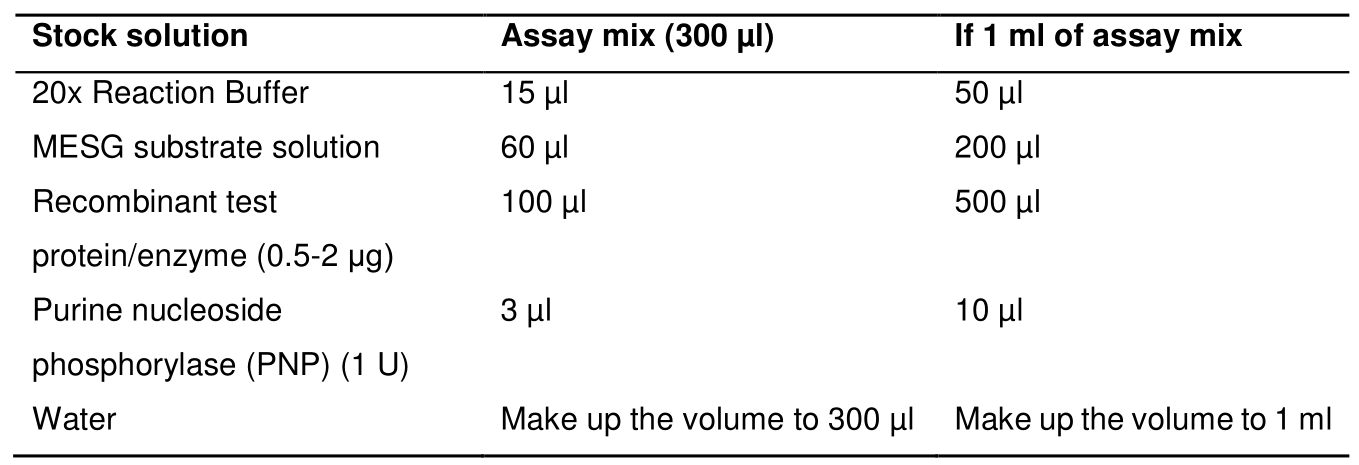
- Prepare blank in a microtiter plate well by adding water instead of recombinant ATPase test protein.
- Prepare the sample volume of recombinant ATPase test protein ranging from 0.5 to 2 µg of protein in water to the total volume to 100 µl.
- Prepare test samples well by adding 100 µl of recombinant ATPase test protein prepared above.
- Prepare positive control microtiter plate well by adding 0.5 units of ATPase (Sigma) instead of recombinant ATPase test protein.
- Incubate the reaction at 22 °C for 10 min in the plate reader.
- Start the reaction by adding 25 µl of ATP (1 mM) as a substrate in each microtiter plate well and mix in the plate reader. If using 1 ml cuvette the amount of ATP needs to be increased accordingly (83 µl).
- Read absorbance in a microtiter plate reader at 360 nm at 0 min and then 10 min intervals for one hour or until saturation point has reached at 22 °C. Incubate the plate in the microtiter plate reader.
- Prepare a 1 mM stock solution of the MESG substrate by adding 20 ml of dH2O directly to the bottle (Component A). Mix extensively to dissolve MESG completely (maybe ~10 min).
Data analysis
- Calculate the ΔA360nm/mintest (ABS360 at saturation point (min) - ABS360 at 0 min for test samples).
- Calculate the ΔA360nm/minblank (ABS360 at saturation point (Min) - ABS360 at 0 min for blank).
- Calculate the ΔΔABS360 = (ΔABS360nm/mintest - ΔABS360nm/minblank). Calculate Average and standard error using all three replicates in each reaction.
- Plot ΔΔABS360 on Y-axis vs recombinant protein concertation on X-axis. The representative data for ΔΔABS360 vs recombinant test protein are shown in Figure 4.
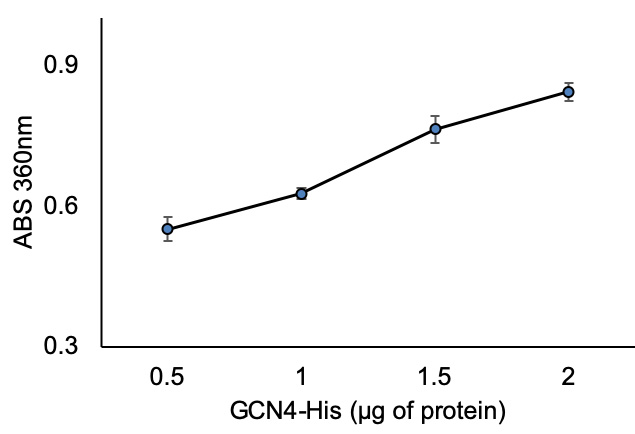
Figure 4. ABS360nm vs. test (GCN4-His) protein concentration - Calculate the amount of inorganic phosphate (Pi) using the phosphate standard curve trendline equation (Figure 3) by substituting y with ΔΔABS360.
- Calculate ATPase activity using the following formula:
ATPase Activity =Pi/(t × V) × D = nmol/min/μl = mU/μl = U/ml
where,
Pi is the inorganic phosphate amount (nmol) from the standard curve (obtained in step 5),
t is the reaction time (min) (time to reach the saturation point),
V is the sample volume (100 µl of recombinant ATPase test protein) added into the reaction well (μl),
D is the sample dilution factor (100 µl/300 µl) = 0.34.
Unit Definition: One unit of ATPase is the amount of enzyme that will generate 1.0 µmol of phosphate per min. - Plot ATPase activity (U/ml) on Y-axis vs recombinant test protein concentration on X-axis.
- To calculate specific activity, divide the U/ml by the amount of protein present in the sample (converted to mg/ml).
Recipes
- 1 M Tris-Cl (pH-8.0)
Tris base12.11 g
Deionized H2O80 ml
Adjust pH to 8.0 with HCl
Make up volume to 100 ml with deionized water
Autoclave at 121 °C for 15 min
Acknowledgments
This work was supported by funds from the Noble Research Institute, LLC. This protocol was derived from Kaundal et al., 2017 manuscript.
Competing interests
The authors declare no financial or non-financial competing interests.
References
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254.
- Brune, M., Hunter, J. L., Corrie, J. E. T. and Webb, M. R. (1994). Direct, Real-Time Measurement of Rapid Inorganic Phosphate Release Using a Novel Fluorescent Probe and Its Application to Actomyosin Subfragment 1 ATPase. Biochemistry 33: 8262-8271.
- Carter, S. G. and Karl, D. W. (1982). Inorganic phosphate assay with malachite green: an improvement and evaluation. J Biochem Biophys Methods 7(1): 7-13.
- Kaundal, A., Ramu, V. S., Oh, S., Lee, S., Pant, B., Lee, H. K., Rojas, C. M., Senthil-Kumar, M. and Mysore, K. S. (2017). GENERAL CONTROL NONREPRESSIBLE4 degrades 14-3-3 and the RIN4 complex to regulate stomatal aperture with implications on Nonhost disease resistance and drought tolerance. Plant Cell 29(9): 2233-2248.
- Rees, D. C., Johnson, E. and Lewinson, O. (2009). ABC transporters: the power to change. Nat Rev Mol Cell Biol 10(3): 218-227.
- Shiue, S. J., Kao, K. M., Leu, W. M., Chen, L. Y., Chan, N. L. and Hu, N. T. (2006). XpsE oligomerization triggered by ATP binding, not hydrolysis, leads to its association with XpsL. EMBO J 25(7): 1426-1435.
- Snider, J., Thibault, G., and Houry, W. A. (2008). The AAA+ superfamily of functionally diverse proteins. Genome Biol 9(4): 216.
- Webb, M. R. (1992). A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc Natl Acad Sci U S A 89(11): 4884-4887.
- Webb, M. R. and Hunter, J. L. (1992). Interaction of GTPase-activating protein with p21ras, measured using a continuous assay for inorganic phosphate release. Biochem J 287 (Pt 2): 555-559.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kaundal, A., Vemanna, R. S. and Mysore, K. S. (2020). The ATPase Activity of Escherichia coli Expressed AAA+-ATPase Protein. Bio-protocol 10(15): e3705. DOI: 10.21769/BioProtoc.3705.
Category
Plant Science > Plant biochemistry > Protein > Activity
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









