- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Contemporaneous Measurement of Outer and Inner Membrane Permeability in Gram-negative Bacteria
(*contributed equally to this work) Published: Vol 10, Iss 5, Mar 5, 2020 DOI: 10.21769/BioProtoc.3548 Views: 7010
Reviewed by: Andrea PuharRon Saar DoverAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Phosphoinositides Coated Beads Binding Assay
Manuel Gálvez-Santisteban [...] Fernando Martin-Belmonte
Feb 5, 2014 12766 Views
Protein-lipid Interaction Analysis by Surface Plasmon Resonance (SPR)
Olga Lucia Baron and David Pauron
Sep 20, 2014 14552 Views

A Fluorescence Dequenching-based Liposome Leakage Assay to Measure Membrane Permeabilization by Pore-forming Proteins
Javier Aguilera [...] Jianjun Sun
May 20, 2021 7108 Views
Abstract
The emergence and rapid spread of multidrug resistance in bacteria have led to the urgent need for novel antibacterial agents. Membrane permeabilization is the mechanism for many antibacterial molecules that are being developed against gram-negative bacteria. Thus, to determine the efficacy of a potential antibacterial molecule, it is important to assess the change in bacterial membrane permeability after treatment. This study describes the protocol for the assays of outer and inner membrane permeability using the fluorescent probes N-phenyl-1-naphthylamine and propidium iodide. Compared with other experiments, such as electron microscopy and the assay of minimal bactericidal concentration, this methodology provides a simpler, faster, and cost-effective way of estimating the membrane-permeabilizing effect and bactericidal efficacy of antibacterial molecules. This study presents an optimized protocol with respect to the classical protocols by incubating bacteria with antibacterial molecules in the culture condition identical to that of antibacterial assays and then detecting the signal of the fluorescent probe in the buffer without broth and antibacterial molecules. This protocol avoids the effect of nutrient deficiency on the physiological status of bacteria and the interference of antibacterial molecules towards the fluorescent probe. Thus, this method can effectively and precisely evaluate the membrane permeability and match the results obtained from other antibacterial assays, such as minimum inhibitory concentration and time–kill curve assays.
Keywords: Gram-negative bacteriaBackground
Multidrug resistance in bacteria is a major public health crisis. Gram-negative bacteria currently pose the greatest threat to public health because of the emergence and rapid spread of carbapenem resistance. Consequently, new antibacterial molecules must be identified to combat this urgent problem. The outer membrane (OM) of gram-negative bacteria is not only the target of several traditional antibiotics to exert antibacterial activities but also of novel therapeutic agents, such as antimicrobial peptides (AMPs). These molecules can disrupt the OM integrity and cause bacterial lysis through permeabilization. Thus, detecting the permeability of the outer and inner membranes is a direct and important means of assessing the efficacy of antibacterial molecules. Electron microscopy is usually applied to observe the morphological changes in OM, and the assay of minimal bactericidal concentration is used to examine bacterial viability after treatment. However, these methods are time-consuming and cannot reflect the real-time change in membrane permeability. In this protocol, we provide a convenient and fast approach for evaluating the antibacterial efficacy of agents with potential membrane-permeabilizing effect by using the fluorescent probes N-phenyl-1-naphthylamine (NPN) and propidium iodide (PI).
NPN is a hydrophobic dye that dissolves sparingly in water with very low fluorescent emission. However, the fluorescence intensity increases sharply when NPN binds with nonpolar substances. The intact OM efficiently blocks NPN out of bacteria to ensure that NPN cannot bind to the hydrophobic tail of phospholipids. By contrast, a strong fluorescence emission can be detected when an OM rupture occurs. Thus, the change in fluorescence intensity of NPN can reflect the efficacy of antibacterial molecules on increasing the permeability of OM. PI is a red-fluorescent nucleic acid stain that can bind to DNA and RNA between the bases. Binding to DNA and RNA leads to an enhancement of PI fluorescence by 20- to 30-fold compared with that in aqueous solutions. Given that PI is a membrane-impermeant stain, it can only label bacteria with a compromised inner membrane (IM). Thus, PI is used in this protocol to determine the change in IM permeability after the treatment of antibacterial molecules.
In most of the related studies, the bacterial OM/IM permeability assay is conducted in a 96-well optical-bottom black plate. Bacteria, fluorescent probe, and antibacterial molecules are mixed together and co-cultured throughout the assay, during which the fluorescence intensity is detected at each time point. To avoid the high background fluorescence emitted by the bacterial culture media, the assay buffer (5 mM HEPES, pH 7.2) is used instead. In this case, bacteria grow under an innutritious condition compared with that of the antibacterial assays such as minimum inhibitory concentration and time–kill curve assays, which may affect the physiological status of bacteria. And some antibacterial molecules may also interfere with the function and performance of the fluorescent probe. As a result, the assay may not exactly reflect the change in permeability caused by the antibacterial molecules. The current work presents an optimized procedure with respect to the previously reported protocols (Ma et al., 2016b; Yarlagadda et al., 2016; Krishnamurthy et al., 2019) by culturing bacteria and exposing them to antibacterial molecules in the same medium as those of other antibacterial assays, harvesting bacteria, and detecting the fluorescent yield in the assay buffer at each time point. This methodology can evaluate the membrane permeability more efficiently and precisely and better match the results obtained from the antibacterial assays.
Materials and Reagents
- Pipette tips
- 1.5 ml Eppendorf tube (Eppendorf Safe-Lock, catalog number: 0 22363204 )
- Steritop 0.22 μm filter unit (Millipore Millex-GP, catalog number: SLGP033RB )
- Flat-bottomed polystyrene 96-well cell culture plates, 0.2 ml well-volume (Corning, Costar®, catalog number: 3599 )
- Glass culture tube 20 mm x 150 mm (sterilized by autoclaving)
- 50 ml glass conical flask (sterilized by autoclaving) (ShuNiu, China)
- E. coli XJ141026 (Isolated from Xijing Hospital)
Note: Other types of E. coli can also be used. - Luria–Bertani (LB) broth (BD/Difco, catalog number: 244620 )
- N-Phenyl-1-naphthylamine (NPN) (Sigma-Aldrich, catalog number: 104043 )
- Propidium iodide (PI (Sigma-Aldrich, catalog number: P4170 )
- Thanatin (synthesized and purified to over 98%)
Note: Thanatin was synthesized with the solid-phase method by applying the Fmoc (9-fluorenylmethyloxycarbonyl) active ester chemistry of as described previously (Fehlbaum et al., 1996). Other AMPs of the experimenter’s choice could also be used instead of thanatin. - Glucose (Sigma-Aldrich, catalog number: G8270 )
- 1 M HEPES solution (Sigma-Aldrich, catalog number: H0887 )
- 10x PBS stock (Life Technologies, Gibco®, catalog number: 70011-044 )
- Acetone (Sigma-Aldrich, catalog number: 650501 )
- Milli-Q filtered water (ddH2O)
- 5 mM HEPES and 5 mM glucose buffer (pH = 7.2) (see Recipes)
Equipment
- Pipettes
- Shaking incubator (Zhicheng, model: ZHWY-200D )
- Static incubators (Taisite Instrument, model: DH4000BII )
- Microplate spectrophotometer (BioTek, model: PowerWave HT )
- Tabletop centrifuge machine (Hanil Science Industrial, model: Smart R17 )
- Fluorescence spectrophotometer (Hitachi, model: F-2500 )
- Water purification system (Millipore, model: Milli-Q Advantage A10 )
- Quartz cuvettes with 1 cm path length (Starna Cells, catalog number: 3-Q-10 )
Software
- Gen 5 (Biotek, USA)
- FL Solutions 2.0 (Hitachi, Japan)
- Excel 2016 (Microsoft, USA)
- Prism 8.0 (GraphPad, USA)
Procedure
- Sample preparation for the membrane permeability assay
- Inoculate a single colony of E. coli XJ141026 into 5 ml LB broth in a glass culture tube and incubate it overnight in a shaking incubator at 220 rpm at 37 °C.
- Take 0.1 ml of stationary growth phase of the E. coli XJ141026 culture and inoculate 10 ml of the LB broth in a 50 ml glass conical flask. Allow the culture to grow for 12 h at 37 °C and 220 rpm.
- Add 100 μl of LB broth to 8 wells of the 96-well microtiter plate. Transfer 100 μl of the culture to the first well and make serial twofold dilutions in the LB broth (final volume of 100 μl per well). Determine the OD600 of each well via the microplate spectrophotometer.
- Calculate the dilution factor according to the above serial OD600 values. Dilute a portion of the remaining culture to OD600 = 0.1 (determined by using the microplate spectrophotometer with a sample volume of 100 μl) in the LB broth to obtain 40 ml diluted bacterial suspension.
- Transfer the diluted bacterial suspension into two 50 ml glass conical flasks, 15 ml per group.
- Add 12 μl of 1,000 μM thanatin (dissolved in sterile PBS) to one of the 15 ml bacterial suspensions, in which the final concentration of thanatin is 0.8 μM. Add an equal volume of sterile PBS (pH = 7.2) to the other flask and use as the untreated control. Mix well and grow at 37 °C and 220 rpm.
- Collect the samples of thanatin-treated and untreated cultures at the different time intervals of 0, 30, 60, 120, 180, and 240 min. For each group, 2 ml culture was collected and separately added into two 1.5 ml Eppendorf tubes, 1 ml per tube.
- Pellet the cells at 8,935 x g for 10 min in 1.5 ml Eppendorf tubes, and remove the supernatant.
- Wash the cells through resuspension in 1 ml of 5 mM HEPES and 5 mM glucose buffer (pH = 7.2) and the pellet cells via centrifugation at 8,935 x g for 5 min. Remove the supernatant.
- Repeat Step A9 twice.
- Resuspend the cells in 1 ml of 5 mM HEPES and 5 mM glucose buffer (pH = 7.2).
- Outer membrane permeability
- To determine the outer membrane permeability of thanatin, add 8 μl of 500 μM NPN (dissolved in acetone) to 1 ml resuspended cells (A11) and vortex well.
- Incubate in darkness for 30 min at room temperature.
- Turn on the fluorescence spectrometer, and allow the lamp to warm up for 30 min.
- Define the operating parameters of the spectrometer as follows: photomultiplier tube (PMT) voltage: 700 V, excitation wavelength: 350 nm, and emission wavelength: 420 nm.
Note: The PMT voltage settings are machine- and sample-dependent. To prevent the fluorescence signal values from exceeding the measurement range, the appropriate PMT voltage for the experiments must be established before a series of experiments is started. To set the appropriate PMT voltage, collect and stain the antimicrobial agent-treated bacteria at the last time point of the experiment, detect the fluorescence signal using different PMT voltages, and choose the optimal PMT voltage that can maximally amplify the signal in the detector range. - Carefully clean a fluorescence cuvette with water and ethanol.
- Transfer 1 ml of the incubated culture to the cuvette and place it in the spectrometer.
- Start the data acquisition and record.
- Inner membrane permeability
- To determine the inner membrane permeability of thanatin, add 5 μl of 1 mM PI (dissolved in sterile ddH2O) to 1 ml of resuspended cells (A11).
- Incubate in darkness for 30 min at room temperature.
- Define the operating parameters of the spectrometer as follows: PMT voltage: 700 V, excitation wavelength: 535 nm, emission wavelength: 617 nm.
- Carefully clean the fluorescence cuvette with water and ethanol.
- Transfer 1 ml of the incubated culture to the cuvette and place it in the spectrometer.
- Start the data acquisition and record.
Data analysis
- Repeat the experiment three times independently.
- Import all the detected values into the GraphPad Prism 8.0 to visualize them graphically. As shown in Figure 1, time and relative fluorescence intensity are depicted in the x- and y-axes, and grouped comparison of untreated control vs. thanatin-treated.
- Use a two-way ANOVA to analyze the data between the different groups.
- OM integrity is damaged by Thanatin in a time-dependent manner, and a distinct increase in the fluorescence intensity of NPN is observed 1 h post-incubation (Figure 1A).
- The increase in fluorescence intensity of PI approximately 2 h post-incubation indicates the increase in IM permeability of the bacteria (Figure 1B).
- The basal fluorescence (Control group in Figure 1) also increases with time, which may due to the increasing number of aging and dead bacteria along with culture time. Thus, the final fluorescence data in the thanatin-treated group are normalized by subtracting the fluorescence values of the corresponding control group at each time point (Figure 2).
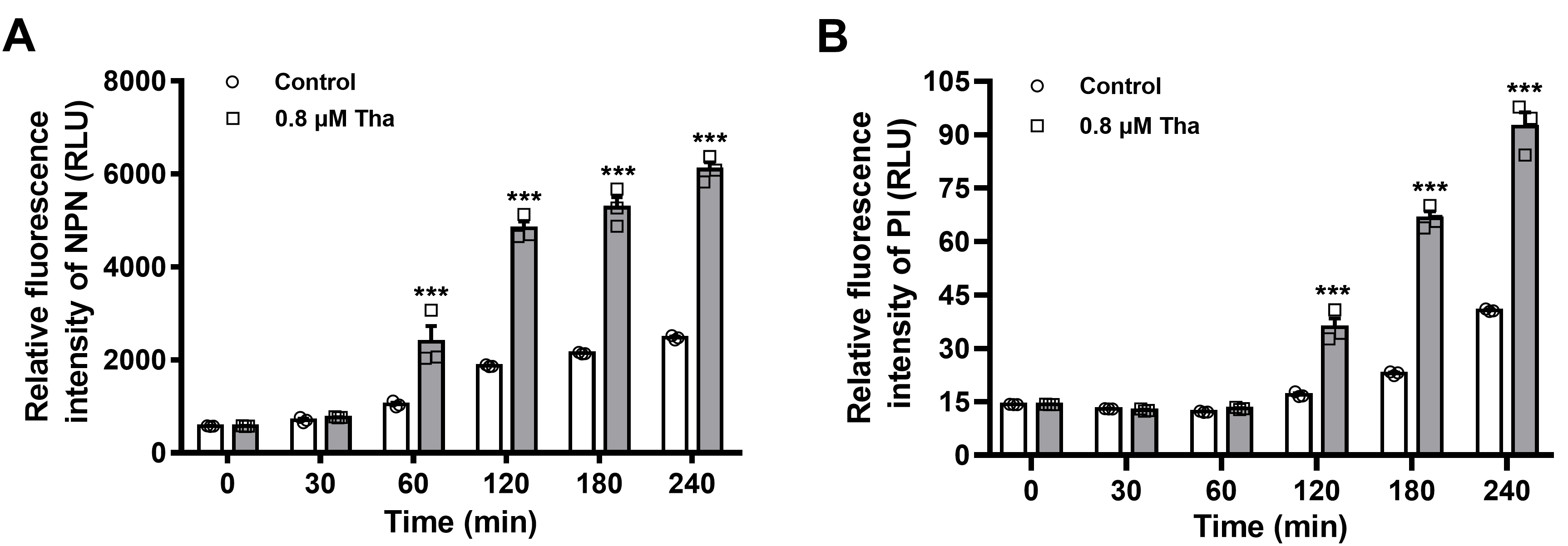
Figure 1. Outer and inner membrane permeabilization of thanatin (Tha) is measured by detecting the fluorescence intensity of NPN (A) and PI (B). All the data are shown as the mean ± SEM of the three independent experiments. P-values were determined via two-way ANOVA; ***P < 0.001 vs. untreated control (Ma et al., 2019).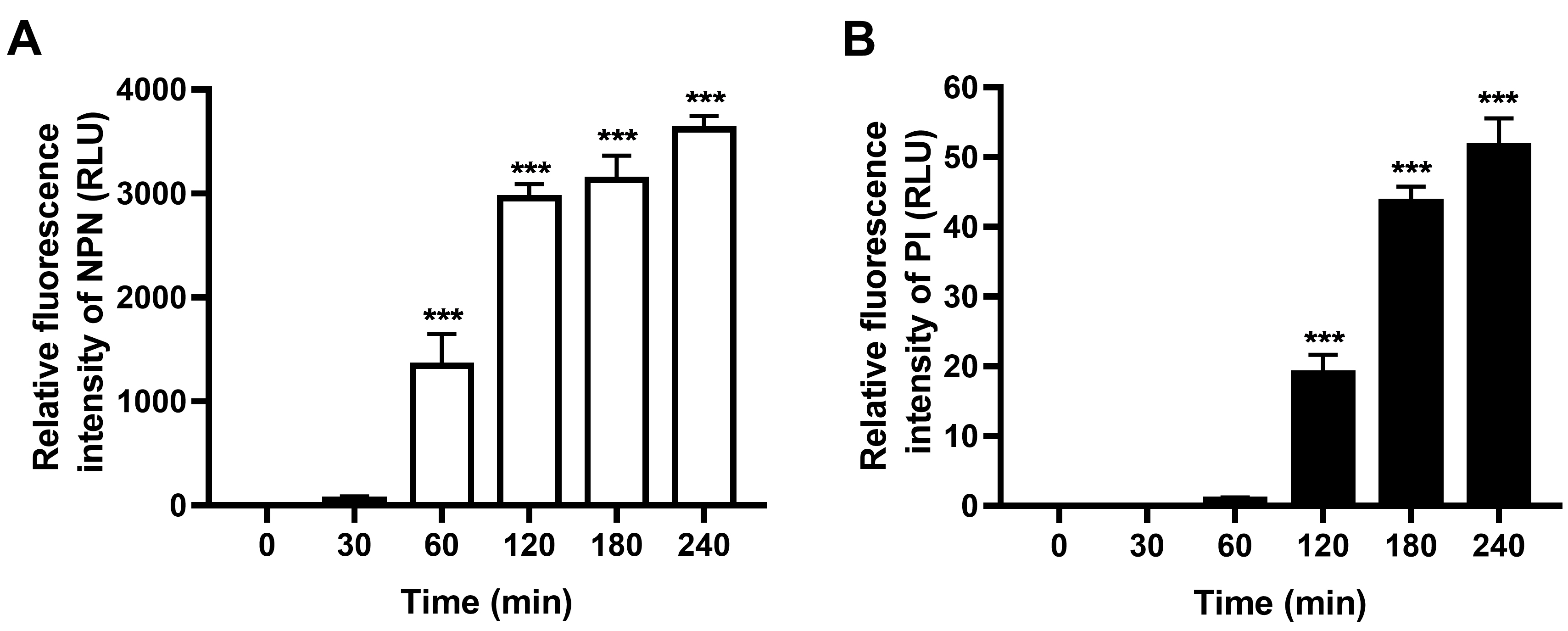
Figure 2. Outer and inner membrane permeabilization of thanatin is measured by detecting the fluorescence intensity of NPN (A) and PI (B) after normalization. All the data are shown as the mean ± SEM of the three independent experiments. P-values were determined via one-way ANOVA; ***P < 0.001.
Recipes
- 5 mM HEPES and 5 mM glucose buffer (pH = 7.2)
1 M HEPES solution is diluted to 5 mM HEPES with ddH2O (pH = 7.2)
Dissolve 90.08 mg glucose in 100 ml of 5 mM HEPES (pH = 7.2)
Sterilize using a 0.22 μm filter
Acknowledgments
This protocol was adapted from previous work (Ma et al., 2016a; Ma et al., 2019). We thank Xiuli Xu and Shan Zhou for providing us with clinical strain from the Department of Clinical Laboratory Medicine of Xijing hospital. This work was supported by grants from the National Natural Science Foundation of China (no. 81673477, 81903671, 81471997 and 81001460).
Competing interests
The authors declare no conflict of interest.
References
- Fehlbaum, P., Bulet, P., Chernysh, S., Briand, J. P., Roussel, J. P., Letellier, L., Hetru, C. and Hoffmann, J. A. (1996). Structure-activity analysis of thanatin, a 21-residue inducible insect defense peptide with sequence homology to frog skin antimicrobial peptides. Proc Natl Acad Sci U S A 93(3): 1221-1225.
- Krishnamurthy, M., Lemmon, M. M., Falcinelli, E. M., Sandy, R. A., Dootz, J. N., Mott, T. M., Rajamani, S., Schaecher, K. E., Duplantier, A. J. and Panchal, R. G. (2019). Enhancing the antibacterial activity of polymyxins using a nonantibiotic drug. Infect Drug Resist 12: 1393-1405.
- Ma, B., Fang, C., Lu, L., Wang, M., Xue, X., Zhou, Y., Li, M., Hu, Y., Luo, X. and Hou, Z. (2019). The antimicrobial peptide thanatin disrupts the bacterial outer membrane and inactivates the NDM-1 metallo-beta-lactamase. Nat Commun 10(1): 3517.
- Ma, B., Niu, C., Zhou, Y., Xue, X., Meng, J., Luo, X. and Hou, Z. (2016a). The disulfide bond of the peptide thanatin is dispensible for its antimicrobial activity in vivo and in vitro. Antimicrob Agents Chemother 60(7): 4283-4289.
- Ma, L., Wang, Y., Wang, M., Tian, Y., Kang, W., Liu, H., Wang, H., Dou, J. and Zhou, C. (2016b). Effective antimicrobial activity of Cbf-14, derived from a cathelin-like domain, against penicillin-resistant bacteria. Biomaterials 87: 32-45.
- Yarlagadda, V., Manjunath, G. B., Sarkar, P., Akkapeddi, P., Paramanandham, K., Shome, B. R., Ravikumar, R. and Haldar, J. (2016). Glycopeptide Antibiotic To Overcome the Intrinsic Resistance of Gram-Negative Bacteria. ACS Infect Dis 2(2): 132-139.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ma, B., Fang, C., Zhang, J., Wang, M., Luo, X. and Hou, Z. (2020). Contemporaneous Measurement of Outer and Inner Membrane Permeability in Gram-negative Bacteria. Bio-protocol 10(5): e3548. DOI: 10.21769/BioProtoc.3548.
Category
Microbiology > Antimicrobial assay > Antibacterial assay
Biochemistry > Lipid > Membrane lipid
Biochemistry > Protein > Interaction > Protein-lipid interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









