- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurement of Sleep and Arousal in Drosophila
Published: Vol 9, Iss 12, Jun 20, 2019 DOI: 10.21769/BioProtoc.3268 Views: 8630
Reviewed by: Adler R. DillmanXiaoliang ZhaoAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measuring Sleep and Activity Patterns in Adult Zebrafish
Fusun Doldur-Balli [...] Allan I. Pack
Jun 20, 2024 1958 Views

Locomotor Activity Monitoring in Mice to Study the Phase Shift of Circadian Rhythms Using ClockLab (Actimetrics)
Andrea Brenna [...] Urs Albrecht
Feb 20, 2025 1773 Views

A Protocol to Assess Time-of-Day-Dependent Learning and Memory in Mice Using the Novel Object Recognition Test
Jordan Mar [...] Isabella Farhy-Tselnicker
Sep 20, 2025 2494 Views
Abstract
Sleep is a conserved neurobehavioral state observed in animals with sufficiently complex nervous systems and is critical for survival. While the exact function of sleep remains unknown, the lack of sleep can have a range of physiological and behavioral effects. Studies in invertebrates and vertebrates have identified conserved neural mechanisms and cellular pathways in control of sleep, wakefulness and arousal. Methodologies to measure sleep have ranged from EEG recordings in humans and rodents to in-depth analysis of locomotor patterns in flies, fish and worms. Here we focus on sleep measurements using activity monitoring in the highly versatile experimental model system, Drosophila melanogaster, which is amenable to a number of genetic, physiological and behavioral manipulations. Further, we also describe methods used to manipulate sleep and wakefulness to understand the neural regulation of sleep and how organisms balance sleep, wakefulness and behavioral arousal. Sleep as a behavioral state is regulated by a number of factors including food, environmental conditions, and genetic background. The methodologies described here provide, a high-throughput approach to study neural regulation of sleep and factors that affect this complex behavior.
Keywords: DrosophilaBackground
Sleep is defined as a state of quiescence that is associated with periods of inactivity, altered brain activity, and heightened sensory threshold. Across species, sleep is tightly regulated by the circadian clock and homeostatic mechanisms, and these processes have been studied extensively in the context of sleep regulation (Hendricks et al., 2000a and 2000b, Shaw et al., 2000, Donlea et al., 2017, Keene and Duboue, 2018, Bringmann, 2019). While the importance of these processes in sleep and wake cycling cannot be overstated, the dynamic regulation of sleep, wakefulness and arousal by other behavioral drives like social engagement and nutritional status have received less attention.
We recently investigated how sleep is regulated in light of competing behavioral drives like courtship and found that sleep is suppressed when the courtship drive is elevated, and courtship behavior is suppressed when sleep drive is increased by deprivation (Aso et al., 2014, Sitaraman et al., 2015a and 2015b, Chen et al., 2017). Here we describe the methods to measure and manipulate sleep amount (by mechanical deprivation and pharmacological treatments) and structure using the high-throughput activity monitoring system that is relatively easy to set-up and comparable to high-resolution video monitoring studies. The sleep recording protocol described here uses the well-established activity monitoring system relying on occlusion of infrared beam by movement of singly housed flies. While a previously published article describes the use of Drosophila activity monitoring system for circadian analysis and sleep, here emphasis is placed on the actual fly rearing, set-up of the activity monitors and circadian analysis (Chiu et al., 2010). Here, we describe the experimental set-up specific to sleep measurements and the detailed analysis of the locomotor data in interpreting sleep amount and architecture. The experimental amenability of the fruit fly system has and will continue to provide key insights into how sleep is influenced by neuronal and environmental factors with careful attention to experimental design, execution, and data interpretation.
Materials and Reagents
- Cheesecloth (Pure Grade 50 100% Unbleached Cotton Cheesecloth Strain, 2 Yards, www.amazon.com)
- Petri dishes (100 x 15 mm, Genesee Scientific, Olympus Plastics, catalog number: 32-107)
- Pipe cleaners (Creativity Street Chenille Stems/Pipe Cleaners 12 Inch x 6 mm, Black from https://www.amazon.com)
- Sleep tubes (TriKinetics Inc., catalog number: PPT5x65 Polycarbonate)
- Rubber bands (Staples)
- Paint brush (Round Pointed Tip Nylon Hair, www.amazon.com)
- Fly tubes and plugs (Genesee Scientific, catalog numbers: 32-116BF and 49-102)
- Paper towel (Kimberly-Clark Kimtech Science Kimwipes, Amazon.com)
- Drosophila (Bloomington Drosophila Resource Center, https://bdsc.indiana.edu)
- Paraffin Wax (Performa Unscented Paraffin Wax Beads from https://www.amazon.com)
- Sucrose (Sigma, catalog number: S8501)
- Agarose (Sigma, catalog number: A9539)
- Dextrose (Sigma, catalog number: G8270-1KG)
- Yeast (Genesee Scientific, catalog number: 62-108)
- Gelidium Agar (Genesee Scientific, catalog number: 66-105)
- Cornmeal (Genesee Scientific, catalog number: 62-100)
- 10% Tegosept (Genesee Scientific, catalog number: 20-258)
- 70% ethanol (Sigma, catalog number: 793213)
- Molasses (Genesee Scientific, catalog number: 62-118)
- Propionic acid (Genesee Scientific, catalog number: 20-271)
- (2-hydroxypropyl)-β-cyclodextrin (Sigma, catalog number: H107)
- Gabaxadol (THIP) (Sigma, catalog number: T101)
- Fly food (see Recipes for Dextrose and Molasses media)
- Fly food with molasses for sick stocks
- Dextrose Fly food for sleep tubes
Equipment
- Thermometer (Easy-Read® thermometer) (Sigma, catalog number: Z425313)
- Stir bar (Fisher Scientific, catalog number: 14-513-83)
- Refrigerator (Samsung from Home Depot or Best buy)
- Paraffin Bath (True Glow by Conair Thermal Paraffin Bath from https://www.amazon.com)
- Drosophila Activity Monitors (Trikinetics)
- Incubator (Percival, DR 36VL)
- Computer (Mac or PC work with binning and analysis software)
- DAM Extension cables (SPLT5: 5-way splitter and CAB10: 10” cables, Trikinetics.com)
- Stereo microscope (Leica M80, Zeiss Stermi 2000, Nikon SMZ745 or any standard stereoscope)
- Light source (Dolan Jenner, KL300 LED or Goose neck)
- CO2 fly pad (Genesee Scientific, catalog number: 59-114)
- CO2 tank and regulator (Airgas)
- Multi-tube Vortex (Troemner, catalog number: 945007/945008)
Software
- DAMFileScan Software (TriKineticsDAMSystem)
- MATLAB (MathWorks, Natick, MA) with signal processing toolbox
- Prism (GraphPad)
Procedure
- Fly stock maintenance
Maintain stocks at 18 °C and 50% relative humidity (RH) under a 12:12 h light/dark cycle in standard fly food vials containing dextrose media (see Fly Food Recipe). Stocks that were sick and not laying enough eggs were transferred to fly media containing molasses (see Fly Food Recipe). Flip stock flies onto fresh food at least once per month. - Fly collection for sleep assays
Collect 32 male flies on a CO2 fly pad under a stereo microscope and place into a standard food vial. Maintain collected flies at 21-25 °C under the same 12:12 h light/dark cycle and 50% RH for at least two days prior to the sleep assay.- Virgin and mated female flies sleep differently and to prevent female flies from laying eggs in the tube we only test virgin female flies. However, this difference can vary depending on genotype and genetic background. Several studies also use sleep tubes containing 5% sucrose and 1% agarose as it prevents egg-laying.
- Use adult flies that are age-matched and in the range of 3-7 days post-eclosion for sleep assays.
- Preparation of Sleep Tubes
- One day before the trial is to be loaded, make sleep tubes to ensure that the food has appropriate moisture and hardness for 3-7 days of sleep recordings (see Recipes). Wrap a rubber band around bundles of 32 sleep tubes; one bundle will be needed per DAM being loaded (can be done in advance, see Figure 1A). While fly media is still hot, pour ~100 ml into each Petri dish. Immediately place bundles of sleep tubes upright into the Sleep Food such that the bundle stands independently with the open ends of the tubes submerged. The submerged portions of the tube will be filled with fly media, which will serve as a food and water source during the experiment. Ensure that tubes are not standing at an angle, as this will result in uneven food levels. Up to seven bundles will fit into each Petri dish. Cover drying sleep tube bundles with cheesecloth.
- When the media in the tube is solidified at room temperature (~30 min), remove the cheesecloth and carefully remove sleep tube bundles from the Sleep Food, ensuring that a plug of agarose remains in each tube. Remove the rubber band and clean the media from the sides of the sleep tubes with a paper towel. Dip the food-containing end of each sleep tube into paraffin wax two to three times to coat this end completely. It is not necessary to wait for the wax to dry between dips, but allow wax to solidify after all dips for approximately 5 s. If wax drips off the end leaving the food exposed, dip again until the food is completely covered. Place waxed tubes onto paper towels to dry at room temperature.
Note: Store dry waxed tubes overnight in a refrigerator in a sealed container with a damp paper towel to maintain humidity. Tubes stored for more than one day may dry out and be unusable. - Remove tubes from the refrigerator at least 2 h before their use to ensure there is no condensation inside and let them equilibrate to room temperature before loading flies.

Figure 1. Preparation and loading of sleep tubes in the activity monitor. A. Sleep tubes (5 in x 65 mm) for loading food/desired nutritional substrate. B. Sleep tubes with flies loaded on the monitor. C. Monitors connected to the power supply unit and computer by 2 or 10” cables in the incubator. D. Food filled tube with paraffin wax seal on one end and a pipe cleaner on the other.
- Loading Activity Monitors
- Cut pipe cleaners into approximately 0.5 cm length, one piece per glass tube to be used.
- Place collected flies onto CO2 pad and use a paint brush to place one fly into the open end of the waxed sleep tube. With the fly inside, cap the open, non-waxed end with a piece of pipe cleaner. Place loaded tubes aside until 32 tubes have been filled.
- Place sleep tubes into the far right and far left columns of the monitor first (see Figure 1B). Push each tube into the holes of the monitor such that the pipe-cleaner end is protruding more than the waxed end.
- Place a rubber band around the top two rows of tubes (see Figure 1B). Do the same for the bottom two rows.
- Place the remaining tubes alternately above and below the rubber band so that they fit snugly and do not move freely (see Figures 1A-1D).
- When all tubes are loaded, align all tubes such that the center of the tube is in the center of the DAM. Align tubes by pressing the loaded DAM onto an even surface, like the tabletop, with the pipe-cleaner side down until the center of all tubes aligns with the center of the monitor. Note that this is the reason that the tubes were pushed further in the pipe cleaner direction in Step D3.
- After loading all DAMs with sleep tubes, place DAMs into a 21 °C incubator set to 50% RH and the desired 12:12 h light/dark cycle the flies were entrained on previously.
- Open DAM Software on the computer and enter monitor range. While you can load up to 120 monitors this is not recommended for sleep experiments as an activity reading has to be made every minute and loading more than 40 monitors can delay reading and cause errors in sleep measurements.
- Plug the monitors into the power supply unit that plugs into the computer that records monitor data as described in (Chiu et al., 2010).
- Ensure that the monitor numbers which are plugged in appear yellow at first pass, then green as they are automatically scanned by the DAMFileScan software.
- Leave monitors undisturbed in the incubator until the conclusion of the trial, changing the temperature if needed. For temperature-sensitive mutations, such as dTRP and Shibire (Kitamoto, 2001; Hamada et al., 2008), typically the temperature will be 21 °C during Day 1, raised to 29 °C at 9:30 AM (lights on) on Day 2, and lowered back to 21 °C at 9:30 AM on Day 3. Typical constant temperature experiments last five days in the incubator at 21 °C or 25 °C.
Data analysis
The locomotor data represented as occlusion of IR beams is output as a .txt file with date and time stamp information. Each raw monitor data file has 32 channels, each channel representing a single sleep tube/fly, and must be converted to single channel files as 1- and 30-min bins for sleep analysis.
- Save and bin data
- Open the DAMFileScanData folder and select monitor numbers to be analyzed (click the first monitor, then hold shift and click the last in order to select all). Copy these files to a new folder named according to the experiment.
- Open DAMFileScan, and click Select Input Data Folder. Select the data folder created in step A1 (see Figure 2).
- Select your “Monitor Range” from lowest to highest monitor number used.
- Set the “Output File Type” to “Channel files,” and set “Extra Readings” to “Sum into bin.”
- Select bin range and length based on the Day-Night light schedule throughout the assay. First select 1 min for Bin Length. Under “First Bin to Save” select the one minute following the time the incubator lights turn on, on the correct starting date for your trial (e.g., if lights turn on at 09:30:00, the first 1-min bin will be 09:31:00). Under Last Bin to Save, select the time the lights turn on during the last day of the trial (e.g., 09:30:00).
- Name the binned files for the first day included in the trial. For example, if Day 1 were July 21, 2017, the file would be named 072117. Click Save.
- The folder named according to the first trial day will appear where the raw monitor files were saved. Immediately rename this folder to include “_1” at the end (e.g., 072117_1).
- Repeat steps 3-5 selecting 30-min for Bin Length. An example screen is shown in Figure 2.
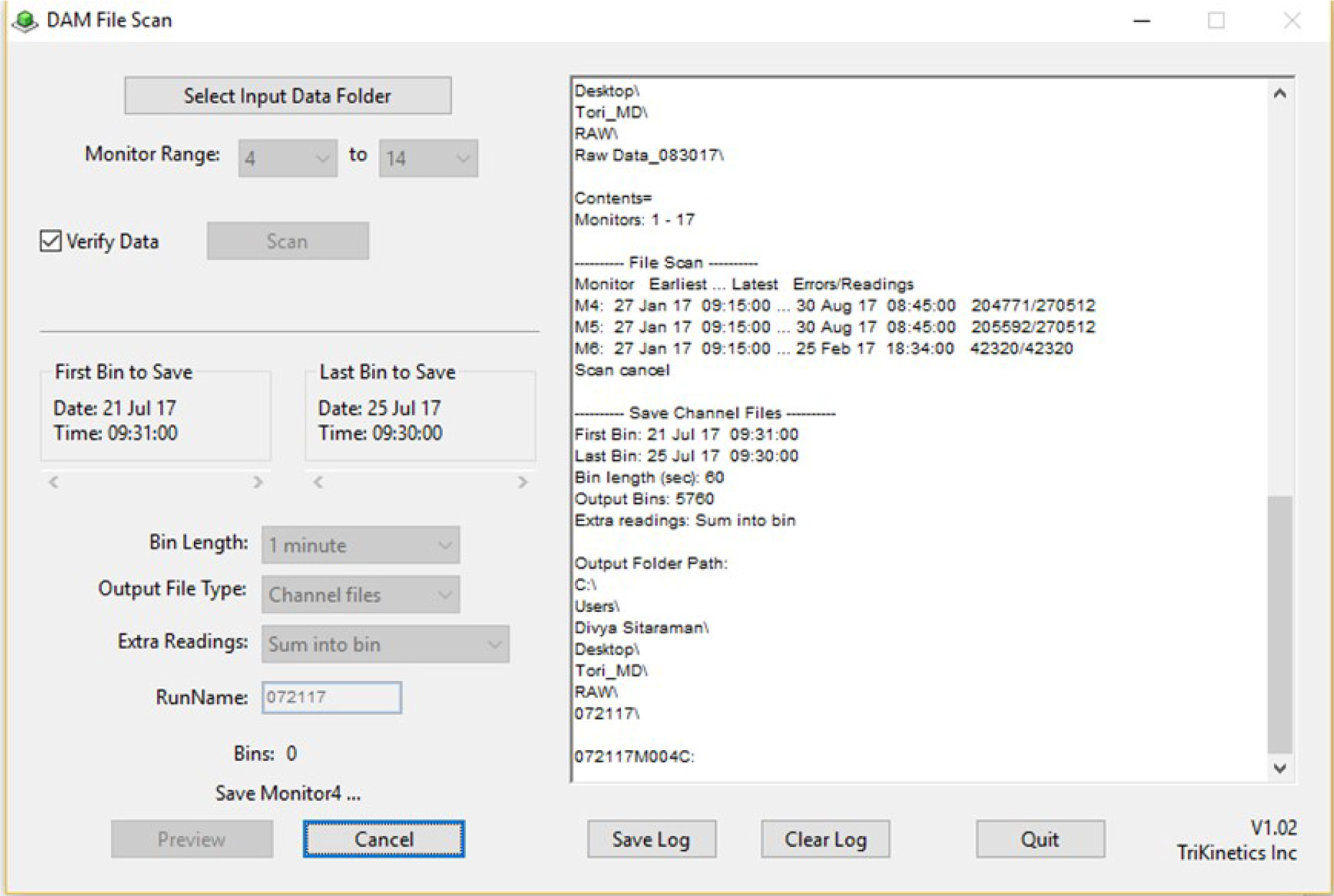
Figure 2. DAM File Scan screen during 1-minute binning. The exemplar experiment ran from July 21, 2017 to July 25, 2017.
- Eliminate channels with dead or immobile flies using MATLAB
- Launch MATLAB, type “scamp” and press Enter. SCAMP program is available here https://www.trikinetics.com/ and was first published in (Donelson et al., 2012).
- Select the folder containing the one-minute bins (e.g., 072118_1) and click “Select Folder”.
- Select the folder containing the thirty-minute bins (e.g., 072118_30) and click “Select Folder”.
- The “Choose boards” page will open the list of monitors that were loaded (Figure 3).
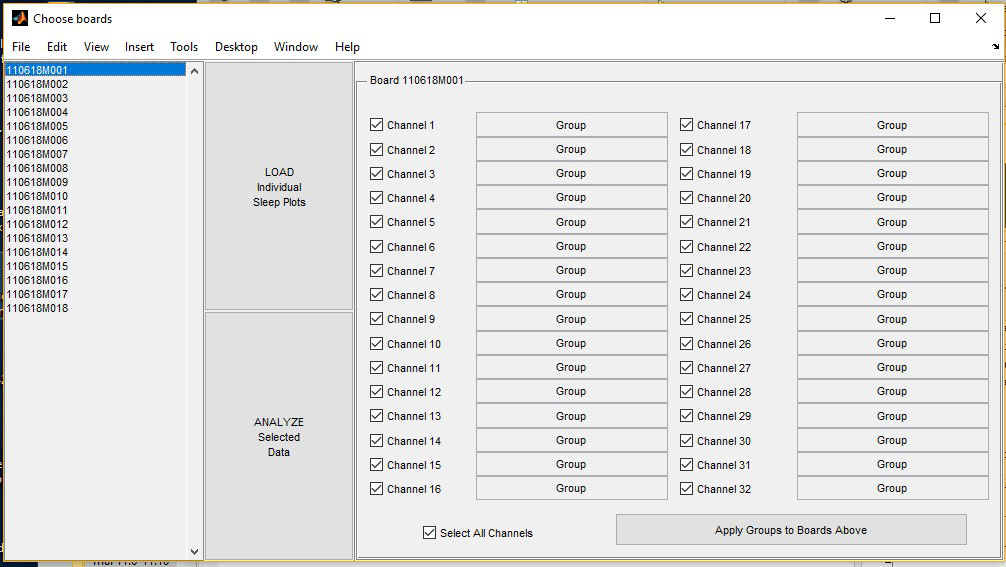
Figure 3. “Choose boards” screen for triage. Exemplar experiment used monitors 1-18 (listed to the left).
- Sleep data analysis post-triage
- Select the first monitor and click “LOAD Individual Sleep Plots” to view each fly’s activity during the trial. This screen display will be used to eliminate dead or immobile flies from future sleep analysis. Ensure that each fly was alive (beam breaks are observed) for the entire length of the trial. Solid blue with no beam breaks indicates that a fly has died. See Figure 4 for example graphs from flies that died during the trial.
- Record the channel number of each fly excluded from analysis (Figure 4).
- Repeat step for all other monitors.
- Navigate to the Choose boards screen (Figure 3) and deselect the box to the left of each channel to be excluded from a given monitor.
- Enter the name of the genotype in each monitor into each of the boxes which say “Group.” This can be done quickly by typing the genotype only once, copying it with Control + C, then continuously holding control while alternating between tapping “Tab” and “V” to paste.
- When all monitors have been sorted for dead and living flies and named for appropriate genotypes, select all monitors for in-depth analysis of sleep characteristics.
- Click “Analyze select data” below the “Individual Sleep Plots” button (see Figure 3).
- A new screen will open- select “Graph Data?” and “Export Data?”. Click “Analyze for chosen bin.”
- Select desired days and click the second button down, “AVERAGE GRAPH and EXPORT”.

Figure 4. Exemplar triage screen. In the above experiment, channels 12, 13, 17, and 25 were excluded. Channel numbers are in numerical order from left to right: the top row contains channels 1-8; the second row contains channels 9-16; the third row contains channels 17-24; and the bottom row contains channels 25-32.
If the trial had a temperature change:- Select only Day 1 and click the “AVERAGE GRAPH and EXPORT” button.
- Excel files will begin populating into the folder where the 1- and 30-min bins were saved. Wait for a screen to open which shows summary preliminary graphs of the data.
- Move all Excel files to a new folder entitled Day 1 Baseline.
- Repeat steps a and b for each subsequent day, naming them accordingly in new folders (for example, Day 2 may be Inhibition or Activation, and Day 3 Recovery).
- Ensure all days are selected and click the “AVERAGE GRAPH and EXPORT” button.
- Excel files will begin populating into the folder where the 1- and 30-min bins were saved. Wait for a screen to open which shows summary preliminary graphs of the data.
- Move all Excel files to a new folder titled Average Data.
- Perform data analysis in Prism or any other graphing or statistical program
Figure 5 shows representative data using GraphPad Prism to plot total sleep (24 h), daytime and nighttime sleep using 12 h Light and 12 h Dark condition.- Open a new file in Prism and create a new XY project for sleep data analysis to plot and perform statistical analysis on different sleep parameters.
- Create Total Sleep Plots.
- For temperature change experiments, open the spreadsheet for the experimental day that follows the 2-3 days of entrainments in 12 h Light and 12 h Dark condition in Microsoft Excel. Total sleep, daytime and nighttime sleep are saved in the Excel file ending with “-stdur.”
- Label the appropriate genotype names from the Excel file column A (see Figure 5).
- Copy and paste the data from the 24 h bin (which will appear in Column D in the Excel file) for each genotype vertically, so that the data will appear in Prism in Column Y under Group A.
- Enter data for all the flies and exclude the calculated average and SEM.
- Repeat Data analysis B and C for each genotype.
- Generate graph by clicking on the Graphs/Data 1 plot (see Figure 5).
- Under “Change Graph Type” select Column.
- Under “Plot”, select Mean with SEM, and click OK.
- Rename axes (for e.g., X-axis are Genotypes and Y-axis is Total Sleep in minutes) and select the “Magic” tool, located bottom right under the “Analysis” toolbar (circled in blue in Figure 6) and select any other graph in order to make them consistent with one another.
- Create Individual Sleep Plots: Sleep profile showing sleep/30 min over a 24 h period.
- Open the Excel document ending in “-s30.” Type the appropriate genotype name from the Excel sheet into Group A in the new Prism sheet. Copy the average and SEM data from the first genotype, and right click to select “Paste transposed.” This will paste the horizontal boxes vertically into Group A in the Prism document.
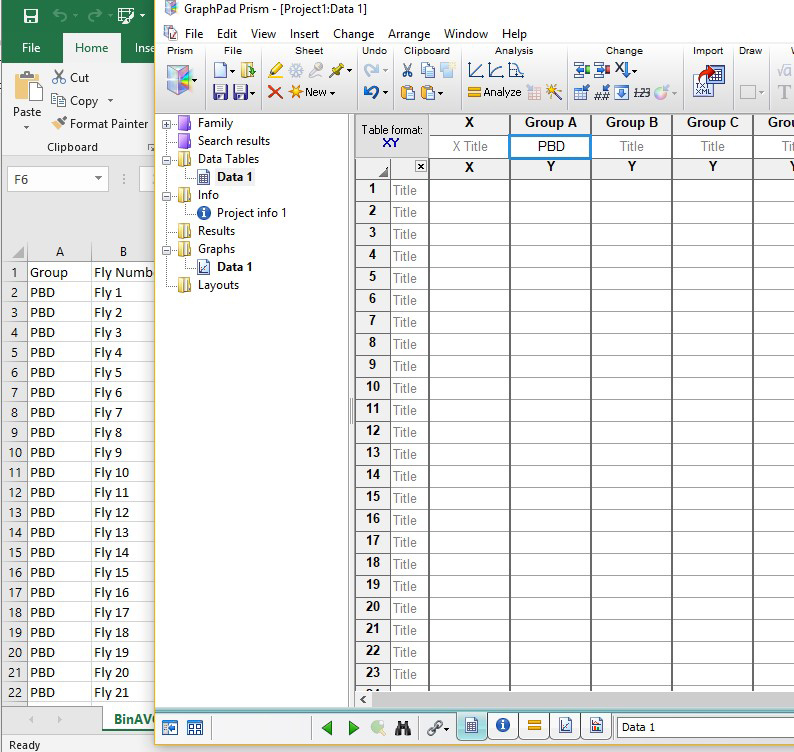
Figure 5. Prism screen (right) and corresponding Excel sheet (left) showing the genotype information and fly number in the activity monitor (1-32) - Click the corresponding graph for the Data Table made in the above step 3a. Select “Grouped” in the drop-down list for “Graph Family” (see Figure 6 for drop-down, “Column” is currently selected). Slide the bar below the Graph Family all the way to the right, and choose the second graph shown, containing a scatter plot with lines between them. Ensure that “Mean with SEM” is selected for “Plot.” Click ok. (Figure 6)
- Open the Excel document ending in “-s30.” Type the appropriate genotype name from the Excel sheet into Group A in the new Prism sheet. Copy the average and SEM data from the first genotype, and right click to select “Paste transposed.” This will paste the horizontal boxes vertically into Group A in the Prism document.
- Create Latency Plots: Indicates time in mins from lights off to start of the first sleep bout (ZT 12).
- Open the Excel document ending in “-latency.” Create a new Prism sheet as above for Total Sleep Plots, and name genotype Groups in Prism as they are listed in Column A of the Excel.
- Copy data from the “2nd 12 h bin” in the Excel (ensuring not to include those points which are labeled as “Average” and “SEM” in Column B) and paste them into the corresponding Group in Prism. Repeat for all genotypes.
- Repeat 2e-2f in Data analysis D to generate the Latency graph.
- Create OA Mean Plots: Indicated number of beam counts/waking minute.
- Open the Excel document ending in “-oamean.” Create a new Prism sheet, and name genotype Groups in Prism as they are listed in Column A of the Excel sheet.
- Copy and paste the data from the “24 h bin” in the Excel document (ensuring not to include those points which are labeled as “Average” and “SEM” in Column B) and paste them into the corresponding Group in Prism.
- Generate the graph as in 2e-2f (under Data analysis D). OA mean is an important measure as it indicated activity of flies when they are awake and not quiescent. In general, flies that sleep a lot show high OA means to balance rest and activity.
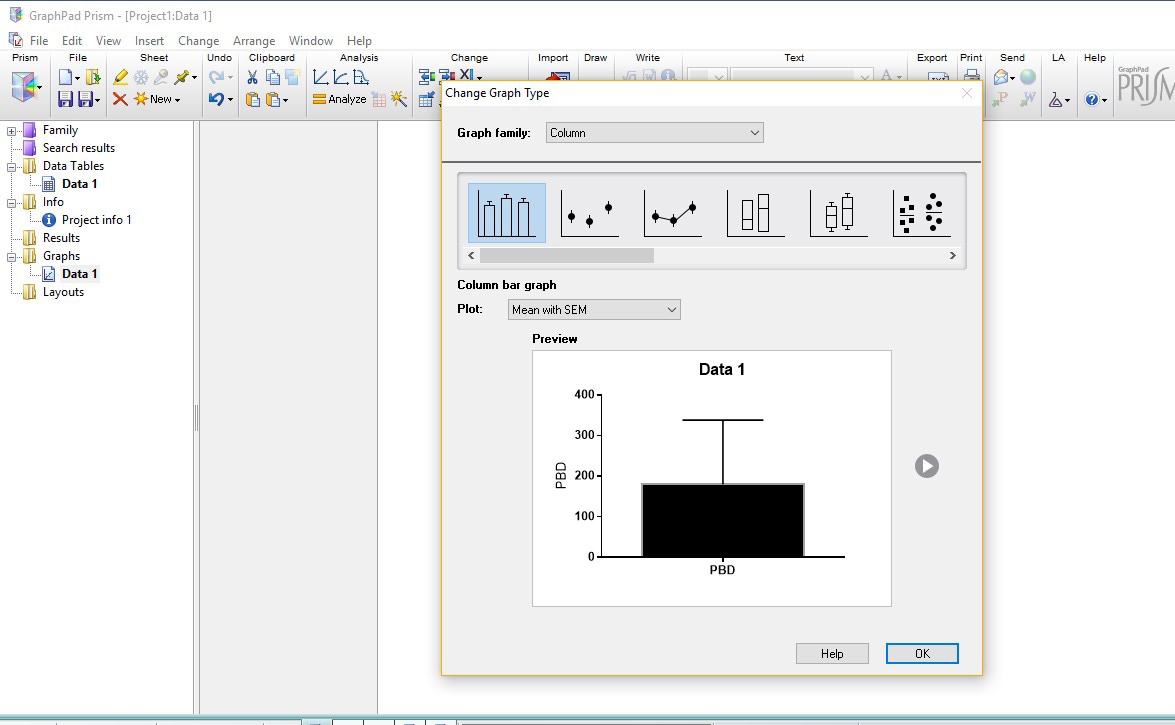
Figure 6. Prism Graph layout screen
- Create Bout Number Plots: Indicates number of sleep bouts during nighttime.
- Open the Excel document ending in “-sfreq.” Create a new Prism sheet, and name genotype Groups in Prism as they are listed in Column A of the Excel sheet.
- Copy and paste the data from the “2nd 12 h bin” in the Excel document (ensuring not to include those points which are labeled as “Average” and “SEM” in Column B) and paste them into the corresponding Group in Prism.
- Generate the graph as in 2e-2f (under Data analysis D). High bout number is an indication of sleep fragmentation.
- Create Mean Bout Length Plots: Indicated average length of each sleep bout.
- Open the Excel document ending in “-smeandur.” Create a new Prism sheet, and name genotype Groups in Prism as they are listed in Column A of the Excel sheet.
- Copy and paste the data from the “2nd 12 h bin” in the Excel document (ensuring not to include those points which are labeled as “Average” and “SEM” in Column B) and paste them into the corresponding Group in Prism.
- Generate the graph as in 2e-2f. Shorter sleep bouts often corresponds with increased number of bouts and is indicative of highly fragmented sleep.
- Example Data
Perform statistical analyses based on the data structure. The above steps extract the features of sleep amount and structure that can be used to understand how specific genetic, behavioral and pharmacological manipulations affects sleep. Figure 7 provides an example figure showing different sleep parameters described above.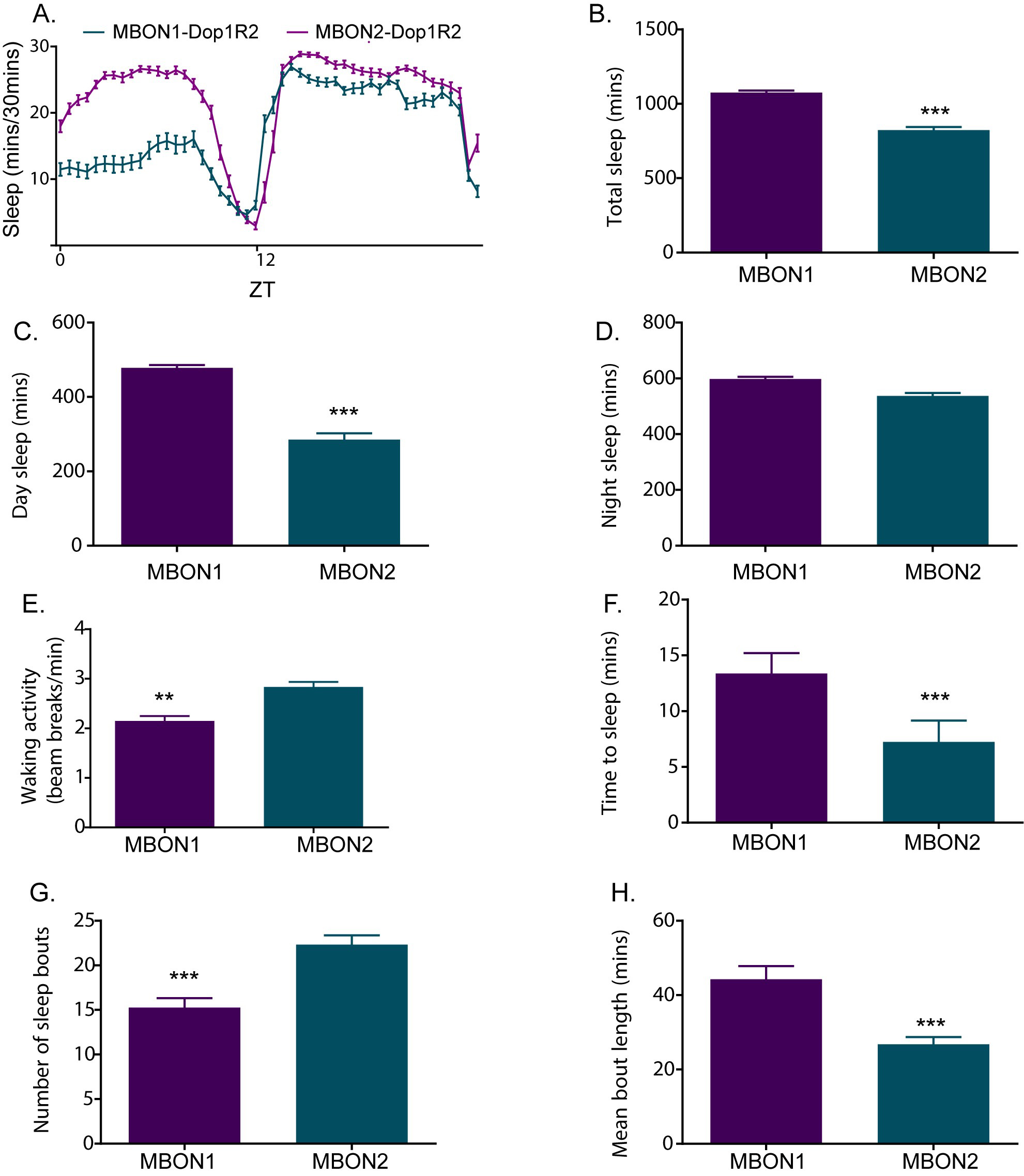
Figure 7. Sleep plots showing different features of sleep quantity and quality. A. Sleep profile of two distinct subsets of neurons (MBON 1 and MBON 2) expressing RNAi (Bloomington Drosophila Resource Center 65997) silencing expression of dopamine receptor Dop1R2. B. Total Sleep in minutes of the two experimental genotypes. C-D. Daytime and nighttime sleep in minutes of the two genotypes. E. Walking activity (beam breaks/minute). F. Latency/time it takes for the flies to start their first sleep bout after ZT (Zeitgeber time) where ZT 12 refers to lights off. G-H. Number of sleep bouts and average bout length of the tested genotypes are indicators of sleep fragmentation. Two genotypes were compared by non-parametric test (unpaired) Mann-Whitney test. Two tailed P-values were used to determine significance (***P < 0.0001 and **P < 0.001).
Sleep manipulation
While genetic approaches to manipulate specific neurons are routinely used in Drosophila to study the neural regulation of sleep, there is an increased focus on measuring neural responses in flies with manipulated sleep drive (Kayser et al., 2015; Beckwith et al., 2017; Chen et al., 2017; Machado et al., 2017). In addition to measuring neuronal responses to sleep manipulation, recent studies have also shown increase in expression of key genes after sleep-deprivation (Toda et al., 2019).
Here we describe methods to increase or decrease sleep drive using mechanical and pharmacological approaches.- Mechanical deprivation: This method entails intermittent shaking of the monitors displacing the flies at random intervals set by the DAM acquisition program. Sleep deprivation can be assessed by measuring sleep during deprivation in the sleep monitors as described above. After loading monitors, flies in the DAM monitor are placed on a horizontal vortexer. The vortexer power cord is attached to a light controller (LC4) that programs the power supply to the unit to deliver pulses that can be set using the DAM acquisition software. The pulse length and frequency can be controlled. We use 20-30 s of shaking per minute throughout the night delivered in pulses lasting 2,3,3,2 s. that are set to Rn setting that stands for random application following a normal distribution. The timing of the pulse, repeat period and distribution can be set at random normal, random uniform and blank/no random setting to increase or decrease the level of deprivation (Figure 8 shows a sample screenshot of the shaker set-up that produces mechanical pulses sufficient to induce sleep-deprivation in wild type CS flies). Flies typically deprived for 16 h show strong sleep rebound. The level of deprivation (sleep lost during deprivation as compared to baseline sleep) and rebound (sleep regained post-deprivation) are variable for different genotypes and genetic background.
- Pharmacological treatments: Alternatively, sleep can be manipulated by feeding flies pharmacological agents to induce arousal or sleep. Carbamazepine (CBZ) is added at a concentration of 0.8 mg/ml to molten food for sleep tubes when it cools to 50-60 °C. Blue or red dye can be added to the food to ensure that flies are ingesting the drug. CBZ is prepared as a stock solution (20 mg/ml) solubilized in 45% (2-hydroxypropyl)-β-cyclodextrin as described in (Agosto et al., 2008). CBZ affects both daytime and nighttime sleep in a manner that depends on the genetic manipulation and genetic background. Nighttime sleep induction can be achieved by treating flies with 0.1 mg/ml Gabaxadol (THIP) as described in Dissel et al., 2015.
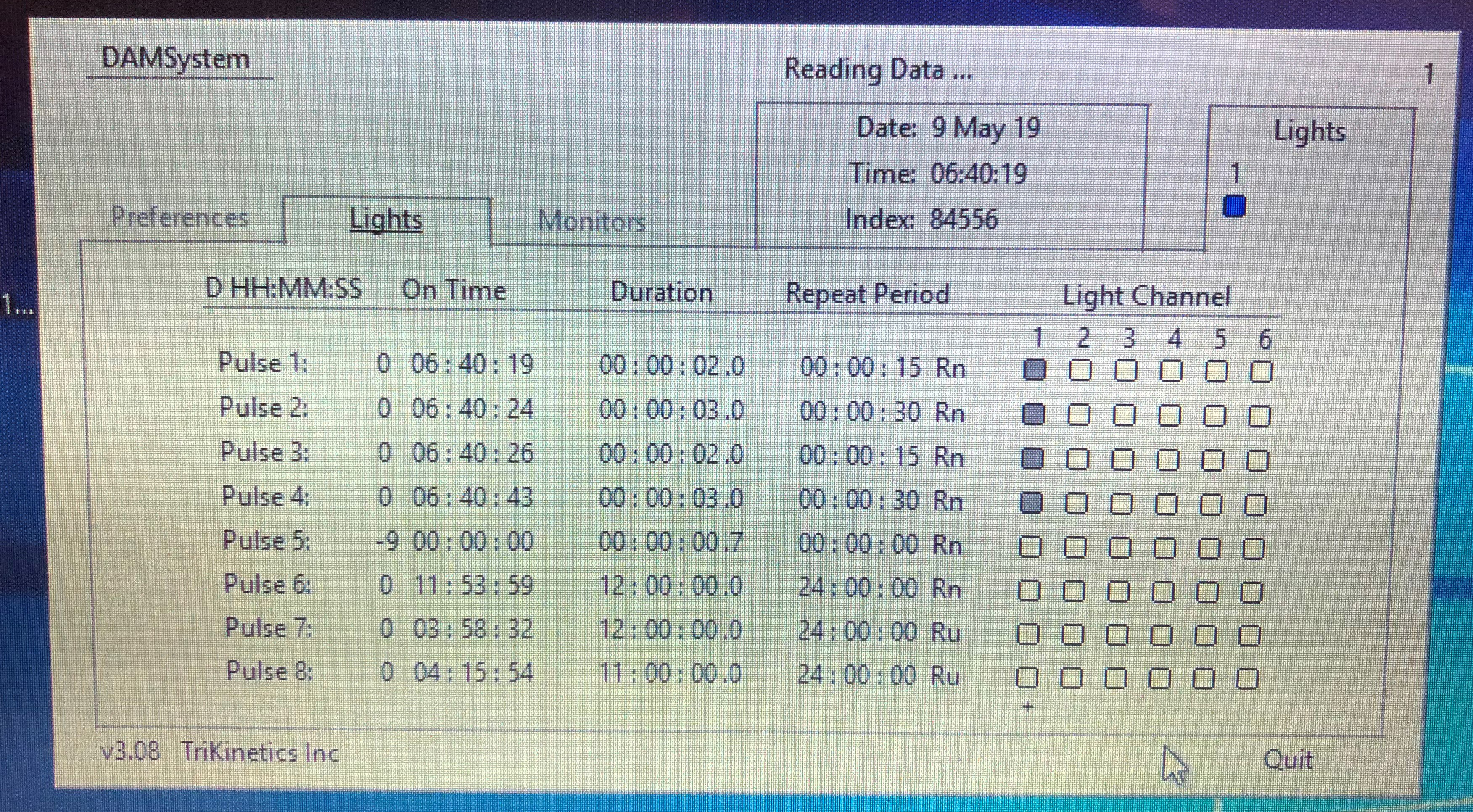
Figure 8. Sample screenshot of the DAM system software controlling mechanical pulses delivered using a Troemner vortexer. The vortexer is directly connected to the light controller that interfaces with the software to control the on time of pulses, duration of pulses and repeat period. - Genetic Deprivation: Several neurons have been implicated in inducing arousal and wakefulness as reviewed in (Donlea et al., 2017). Nighttime activation of subsets of dopamine system neurons (labeled by broad dopamine drivers like Th-Gal4 or more targeted drivers like 54B-Gal4 as shown in Figure 9) and octopamine system neurons (targeted by Tdc2-Gal4) by mis-expression of dTrpA1, a temperature gated non-specific cation channel, induces increased wakefulness when the temperature is raised from 21 to 28-30 °C (Kume et al., 2005; Crocker and Sehgal, 2008; Seidner et al., 2015, Sitaramanet al., 2015a and 2015b). Unlike mechanical deprivation we find that activation of these neurons does not always result in sleep rebound post deprivation when the temperature is lowered to 21 °C.
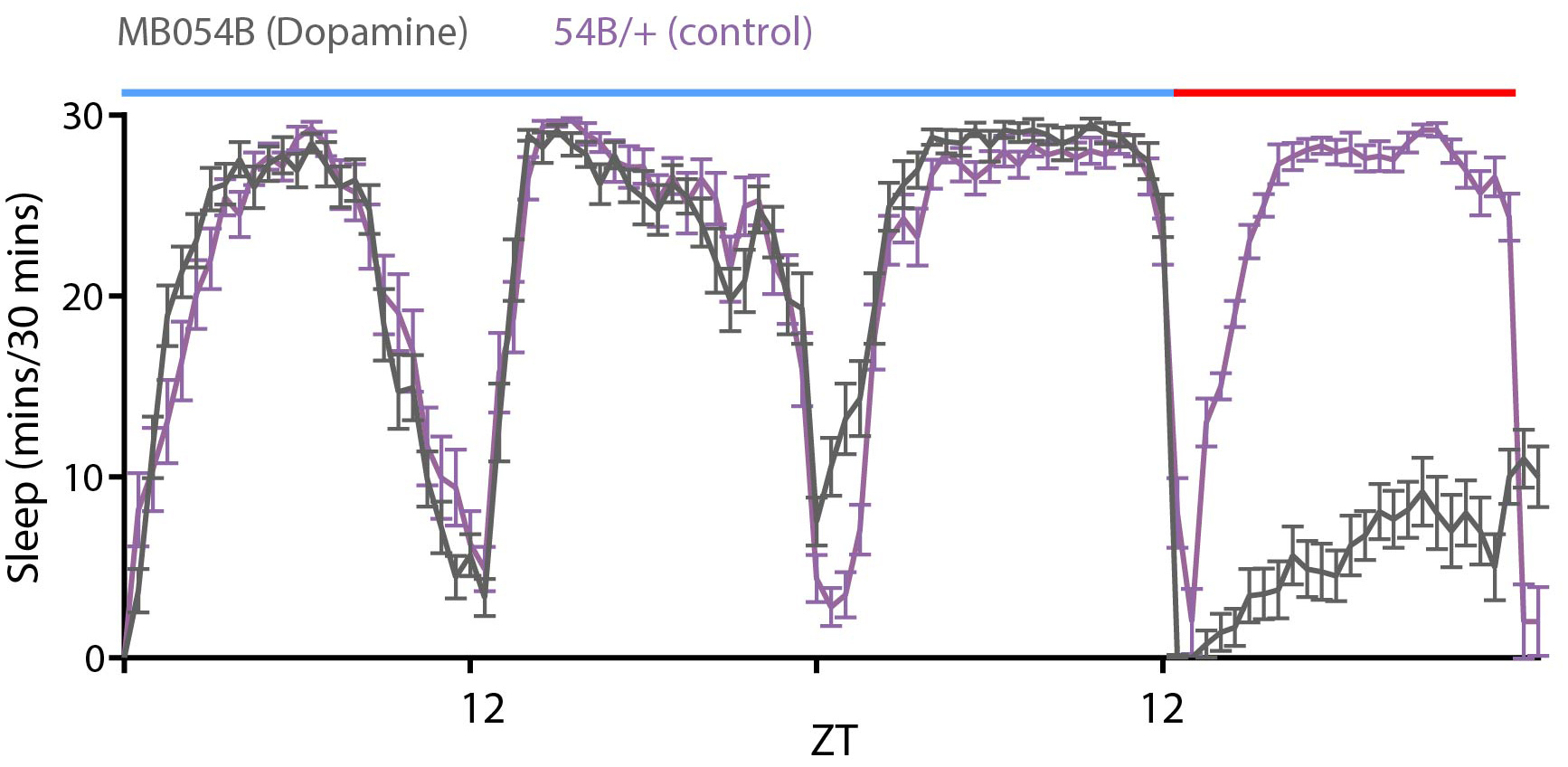
Figure 9. Sleep profile of flies expressing dTrpA1, a temperature-sensitive ion channel in PAM dopamine neurons labeled by MB054B (grey). Control flies (purple) with the MB054B transgene without dTrpA1 expression show no deprivation when the temperature was raised from 21°C (blue bar) to 28 °C (red bar).
Recipes
- Fly food with molasses for sick stocks (1.5 L)
105 g corn meal
44 g yeast
12 g agar
100 ml molasses
1.4 L tap water
16 ml of 10% Tegosept in 70% ethanol
Note: Heat and stir mixture of all ingredients except Tegosept until the mixture reaches 90 °C. Remove from heat and allow to cool for 20 min. Add 10% Tegosept in 70% ethanol, stir, and allow to cool for 40 more min. Pour into fly tubes and cover with cheesecloth to dry overnight. Cap food with foam plugs and store in refrigerator. - Dextrose Fly food for sleep tubes (1 L)
48 g corn meal
8 g agar
25 g yeast
100 g dextrose (D-glucose)
16 ml of 10% Tegosept in 70% ethanol
1.8 ml Propionic acid
Note: Remove some water to hydrate cornmeal and yeast. Stir in dextrose and agar. Turn on heat and bring to boil. Add cornmeal-yeast mixture and boil for another 15-20 min. Turn off the heat and add Tegosept and propionic acid when the food is cool.
Acknowledgments
This work was supported by NIH Grant # 1R15GM125073-01 to DS.
Competing interests
The authors declare no competing financial interests.
References
- Agosto, J., Choi, J. C., Parisky, K. M., Stilwell, G., Rosbash, M. and Griffith, L. C. (2008). Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci 11(3): 354-359.
- Aso, Y., Sitaraman, D., Ichinose, T., Kaun, K. R., Vogt, K., Belliart-Guerin, G., Placais, P. Y., Robie, A. A., Yamagata, N., Schnaitmann, C., Rowell, W. J., Johnston, R. M., Ngo, T. T., Chen, N., Korff, W., Nitabach, M. N., Heberlein, U., Preat, T., Branson, K. M., Tanimoto, H. and Rubin, G. M. (2014). Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. Elife 3: e04580.
- Bringmann, H. (2019). Genetic sleep deprivation: using sleep mutants to study sleep functions. EMBO Rep 20(3).
- Chen, D., Sitaraman, D., Chen, N., Jin, X., Han, C., Chen, J., Sun, M., Baker, B. S., Nitabach, M. N. and Pan, Y. (2017). Genetic and neuronal mechanisms governing the sex-specific interaction between sleep and sexual behaviors in Drosophila. Nat Commun 8(1): 154.
- Chiu, J. C., Low, K. H., Pike, D. H., Yildirim, E. and Edery, I. (2010). Assaying locomotor activity to study circadian rhythms and sleep parameters in Drosophila. J Vis Exp(43).
- Crocker, A. and Sehgal, A. (2008). Octopamine regulates sleep in Drosophila through protein kinase A-dependent mechanisms. J Neurosci 28(38): 9377-9385.
- Dissel, S., Angadi, V., Kirszenblat, L., Suzuki, Y., Donlea, J., Klose, M., Koch, Z., English, D., Winsky-Sommerer, R., van Swinderen, B. and Shaw, P. J. (2015). Sleep restores behavioral plasticity to Drosophila mutants. Curr Biol 25(10): 1270-1281.
- Donelson, N. C., Kim, E. Z., Slawson, J. B., Vecsey, C. G., Huber, R. and Griffith, L. C. (2012). High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the "tracker" program. PLoS One 7(5): e37250.
- Donlea, J. M., Alam, M. N. and Szymusiak, R. (2017). Neuronal substrates of sleep homeostasis; lessons from flies, rats and mice. Curr Opin Neurobiol 44: 228-235.
- Hamada, F. N., Rosenzweig, M., Kang, K., Pulver, S. R., Ghezzi, A., Jegla, T. J. and Garrity, P. A. (2008). An internal thermal sensor controlling temperature preference in Drosophila. Nature 454(7201): 217-220.
- Hendricks, J. C., Finn, S. M., Panckeri, K. A., Chavkin, J., Williams, J. A., Sehgal, A. and Pack, A. I. (2000a). Rest in Drosophila is a sleep-like state. Neuron 25(1): 129-138.
- Hendricks, J. C., Sehgal, A. and Pack, A. I. (2000b). The need for a simple animal model to understand sleep. Prog Neurobiol 61(4): 339-351.
- Keene, A. C. and Duboue, E. R. (2018). The origins and evolution of sleep. J Exp Biol 221(Pt 11).
- Kitamoto, T. (2001). Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol 47(2): 81-92.
- Kume, K., Kume, S., Park, S. K., Hirsh, J. and Jackson, F. R. (2005). Dopamine is a regulator of arousal in the fruit fly. J Neurosci 25(32): 7377-7384.
- Machado, D. R., Afonso, D. J., Kenny, A. R., Oztu Rk-Colak, A., Moscato, E. H., Mainwaring, B., Kayser, M. and Koh, K. (2017). Identification of octopaminergic neurons that modulate sleep suppression by male sex drive. Elife 6: e23130.
- Seidner, G., Robinson, J. E., Wu, M., Worden, K., Masek, P., Roberts, S. W., Keene, A. C. and Joiner, W. J. (2015). Identification of neurons with a privileged role in sleep homeostasis in Drosophila melanogaster. Curr Biol 25(22): 2928-2938.
- Shaw, P. J., Cirelli, C., Greenspan, R. J. and Tononi, G. (2000). Correlates of sleep and waking in Drosophila melanogaster. Science 287(5459): 1834-1837.
- Sitaraman, D., Aso, Y., Jin, X., Chen, N., Felix, M., Rubin, G. M. and Nitabach, M. N. (2015a). Propagation of homeostatic sleep signals by segregated synaptic microcircuits of the Drosophila mushroom body. Curr Biol 25(22): 2915-2927.
- Sitaraman, D., Aso, Y., Rubin, G. M. and Nitabach, M. N. (2015b). Control of sleep by dopaminergic inputs to the Drosophila mushroom body. Front Neural Circuits 9: 73.
- Toda, H., Williams, J. A., Gulledge, M. and Sehgal, A. (2019). A sleep-inducing gene, nemuri, links sleep and immune function in Drosophila. Science 363(6426): 509-515.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Driscoll, M. E., Hyland, C. and Sitaraman, D. (2019). Measurement of Sleep and Arousal in Drosophila. Bio-protocol 9(12): e3268. DOI: 10.21769/BioProtoc.3268.
Category
Neuroscience > Behavioral neuroscience > Sleep and arousal
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








