- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Microbial Mutagenicity Assay: Ames Test
Published: Vol 8, Iss 6, Mar 20, 2018 DOI: 10.21769/BioProtoc.2763 Views: 36154
Reviewed by: Modesto Redrejo-RodriguezAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Stationary-phase Mutagenesis Soft-agar Overlay Assays in Bacillus subtilis
Karla Viridiana Castro-Cerritos [...] Mario Pedraza-Reyes
Dec 5, 2017 7411 Views
Multiple Stepwise Gene Knockout Using CRISPR/Cas9 in Escherichia coli
Enrico König [...] Guido Grandi
Jan 20, 2018 28643 Views

Comprehensive Mapping of EZ-Tn5 Transposon Insertion Sites in Pseudomonas argentinensis SA190 Using RATE-PCR
Büsra Elkatmis [...] Maged M. Saad
Jul 20, 2025 1750 Views
Abstract
The Microbial mutagenicity Ames test is a bacterial bioassay accomplished in vitro to evaluate the mutagenicity of various environmental carcinogens and toxins. While Ames test is used to identify the revert mutations which are present in strains, it can also be used to detect the mutagenicity of environmental samples such as drugs, dyes, reagents, cosmetics, waste water, pesticides and other substances which are easily solubilized in a liquid suspension. We present the protocol for conducting Ames test in the laboratory.
Keywords: MutagenicityBackground
The Microbial Ames test is a simple, rapid and robust bacterial assay consisting of different strains and applications of Salmonella typhimurium/E. coli, used for ascertaining the mutagenic potential (Levin et al., 1982; Gupta et al., 2009). In 1975, Ames and his followers standardized the traditional Ames assay protocol and reappraised in 1980’s (Maron and Ames, 1983). Induction of new mutations replacing existing mutations allows restoring of gene function. The newly formed mutant cells are allowed to grow in the absence of histidine and form colonies, hence this test is also called as ‘Reversion assay’ (Ames, 1971). While traditional Ames test is quite laborious and time consuming for initial monitoring of mutagenic compounds, miniaturization of liquid suspension significantly impacted the usability by making it more convenient. The standard doses (2 µl, 5 µl, 10 µl, 50 µl and 100 µl) were set to evaluate the mutagenicity from lower to higher concentration (Hayes, 1982). Mice liver has been used as a tissue for preparing homogenate 9,000 x g (S9 hepatic fraction) whereas in S9 mix, hepatocytes are used to minimize the mammalian metabolic activation formed in the mice liver. In Ames bioassay, the sensitivity of a compound for mutagenicity is based on the knowledge that a substance which is mutagenic in the presence of liver enzymes metabolizing compound might be a carcinogen (Mathur et al., 2005).
Genetic Approach: The Salmonella/E. coli tester strains: Several strains of Salmonella typhimurium have been used in Ames assay which requires histidine synthesis to assess the mutagenicity. In the histidine operon, each tester strain contains a different mutation. In addition to the histidine mutation, the standard tester strain of Salmonella typhimurium contains other mutations that greatly enhance their ability to detect the mutations (Figure 1). One of the mutations (rfa) causes partial loss of the lipopolysaccharides barrier that coats the surface of the bacteria and increases permeability to large molecules such as benzo[a]pyrene allowing not to penetrate in the normal cell wall (Mortelman and Zeiger, 2000). The mutagens present in the tested samples give rise to induced revertants on a minimal medium (absence of histidine). They are further used to observe revertants in previously mutated strains (that are not able to grow in a medium without histidine). The other mutation (uvrB) is a deletion mutation in which deletion of a gene, coding for the DNA excision repair system, causing gradually increased sensitivity in detecting many mutagens (Ames et al., 1973a). The reason behind this mutation is the deletion excising the uvrB gene emulsifying these bacteria requiring biotin for growth. The standard strains such as TA 97, TA 98, TA 100 and TA 102 contain the R-factor plasmid, pKM101. These R-factor strains are reverted by a number of mutagens that are detected weakly or not at all with the non R-factor parent strains (Ames et al., 1975a).
Figure 1. Genetic approach for assessing the mutagenicity in Salmonella strains (modified from https://en.wikipedia.org/wiki/Ames_test)
Many studies (Ames et al., 1975b; Levin et al., 1982) revealed that development of plasmid pKM101 in TA 1535 and TA 1538 strains leads to complement other isogenic strains such as TA 98, TA 100, TA 104 and TA 102. The his G46 mutation in TA 100 and TA 1535 codes for the first enzyme of histidine biosynthesis (hisG) (Ames et al., 1975b). This mutation, determined by DNA sequence analysis, substitutes proline (-GGG-) for leucine (-GAG-) in the wild type organism (Barnes et al., 1982). The tester strains TA 1535 and its R-factor derivative present in TA 100, detect mutagens which causes base-pair substitutions generally at one of these G-C pairs. The hisD3052 mutation in TA 1538 and TA 98 is in the hisD gene coding for histodinol dehydrogenase. TA 1538 and its R-factor derivative TA 98 detect various frameshift mutagens in repetitive sequences as ‘hot spots’ resulting in a frame shift mutation (Walker and Dobson, 1979; Shanabruch and Walker, 1980) (Table 1).
Table 1. Genotype of the Salmonella strain used for mutagenesis testing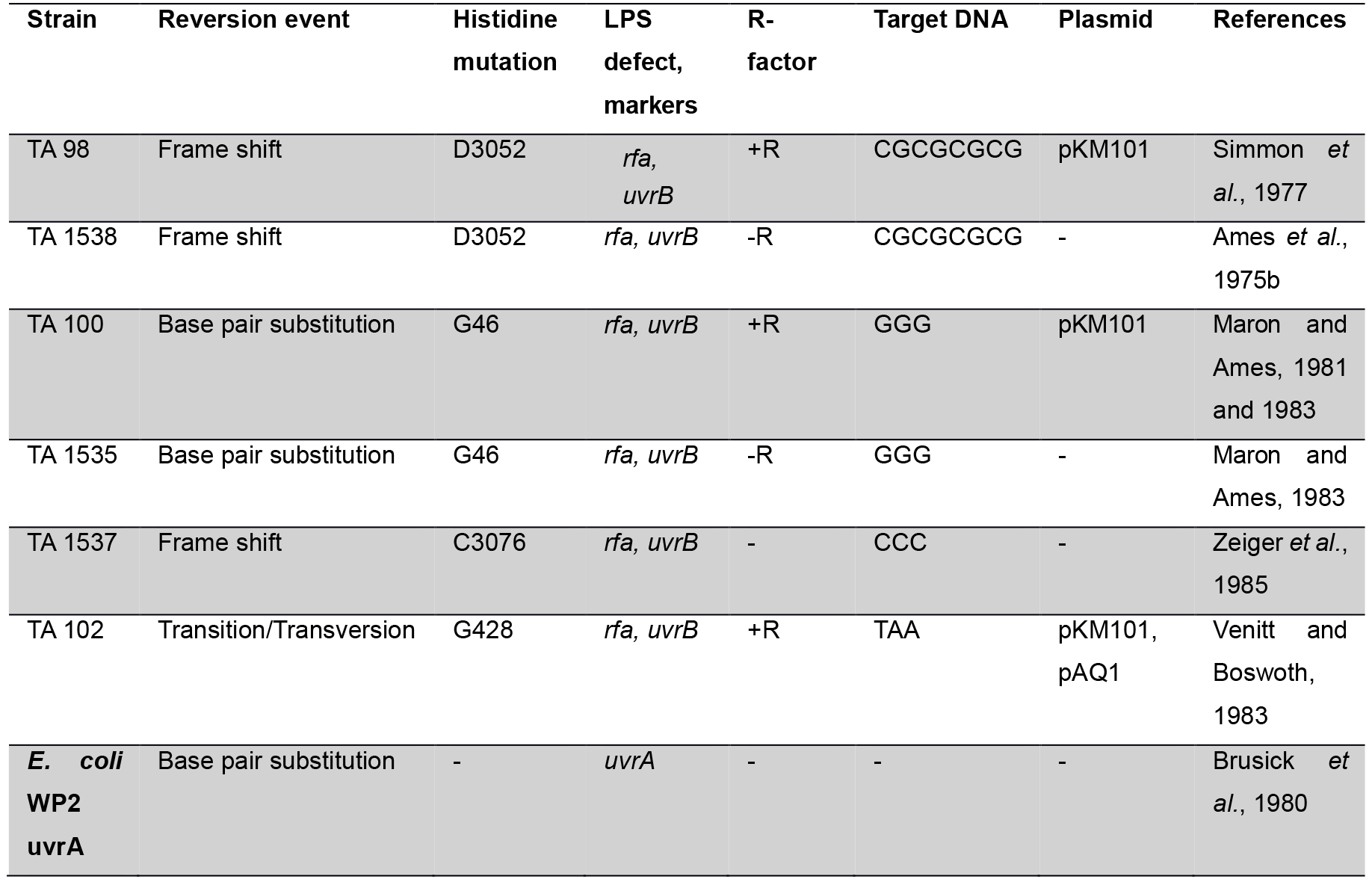
Levin et al. (1982) described a standard strain Salmonella typhimurium bacterium called TA 102 which was used to evaluate the effect of some compounds reacting with nucleotides AT. Tester strain TA102 containing nucleotides AT, present in hisG gene carrying plasmid pAQ1. There are certain mutagenic agents which are detected by TA 102 but not by TA 1535, TA 1537, TA 1538, TA 98 and TA 100 (Wilcox et al., 1990). Before performing experiment, a new set of fresh strains are prepared; and the genotypes are assessed (R-factor, His, rfa and uvrB mutations). For these, we refer readers to many excellent reviews (Walker, 1979; Czyz et al., 2002; Fluckiger-Isler et al., 2004).
Certain carcinogens present in active forms in biological reaction are easily catalyzed by cytochrome-P450. Metabolic activation system is absent in Salmonella, and in order to improve the potentiality of bacterial test systems, liver extracts of Swiss albino mice are used. This serves as a rich source in converting carcinogens to electrophilic chemicals that are incorporated to detect in vivo mutagens and carcinogens (Garner et al., 1972; Ames et al., 1973a). The crude liver homogenate as 9,000 x g S9 fraction contains free endoplasmic reticulum, microsomes, soluble enzymes and some cofactors set with S9 concentration to 10% (Franz and Malling, 1975). The oxygenase requires the reduced form of Nicotinamide Adenine Dinucleotide Phosphate (NADP) which is generally in situ by the action of glucose-6-phosphate dehydrogenase and reducing NADP both work as cofactors in assay (Prival et al., 1984; Henderson et al., 2000). While water is considered as a negative control, sodium azide, 2-nitrofluorine and mitomycin for TA 98, TA 100 and TA 102 without S9 metabolic activation and 2-anthramine with S9 hepatic fraction are used as positive controls for conducting the test (Table 2). Before performing the experiment, fresh solutions must be prepared.
Table 2. Positive controls with and without S9 metabolic activation (DeFlora et al., 1984)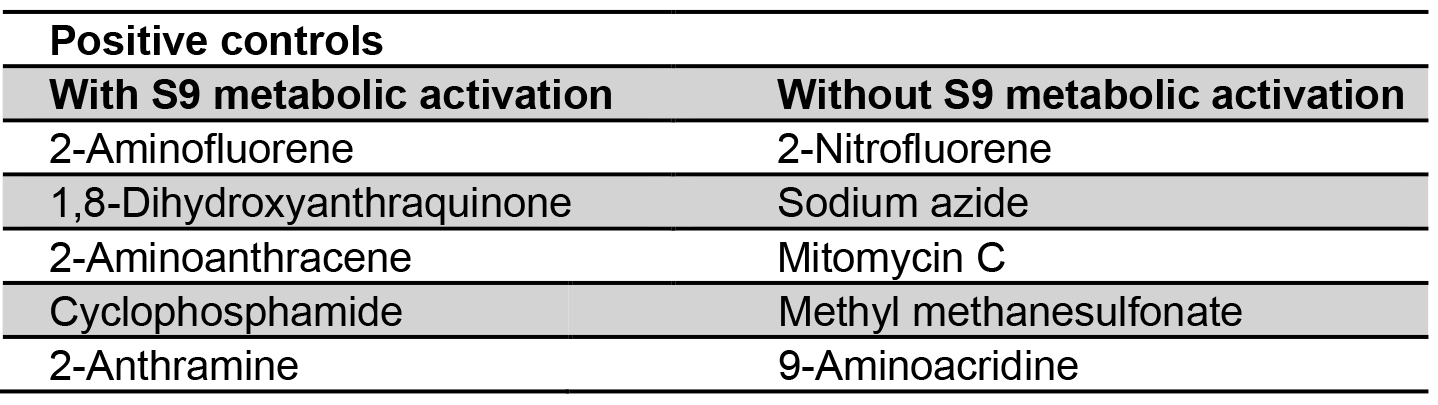
Spontaneous Reversion Control: Each strain of Salmonella contains a specific mutant range. Selection of solvents shows the effect on the frequency range of spontaneous mutant (Maron and Ames, 1983) (Table 3). The range of revertants varies in research laboratories. The spontaneous revertants are visible through unaided eyes (Figure 2).
Table 3. Spontaneous revertants control values for various strain types and number of revertants (Mortelmans and Stocker, 1979)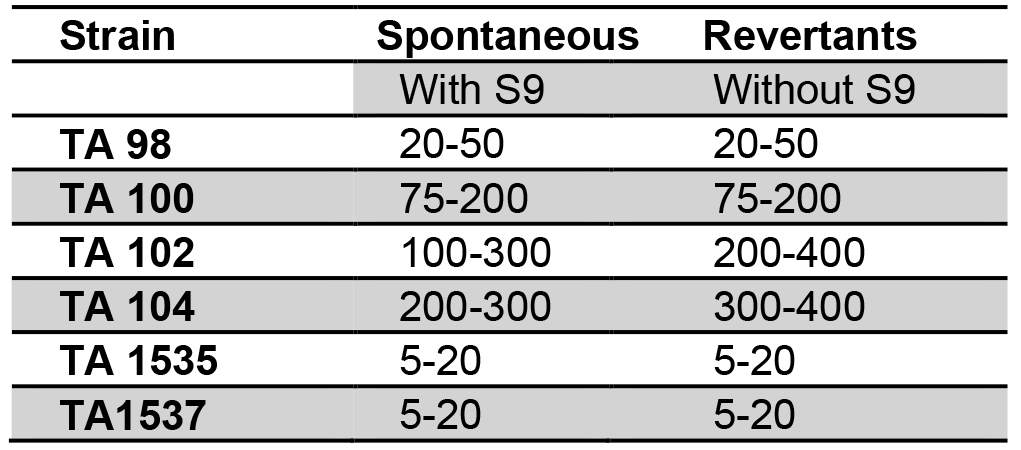
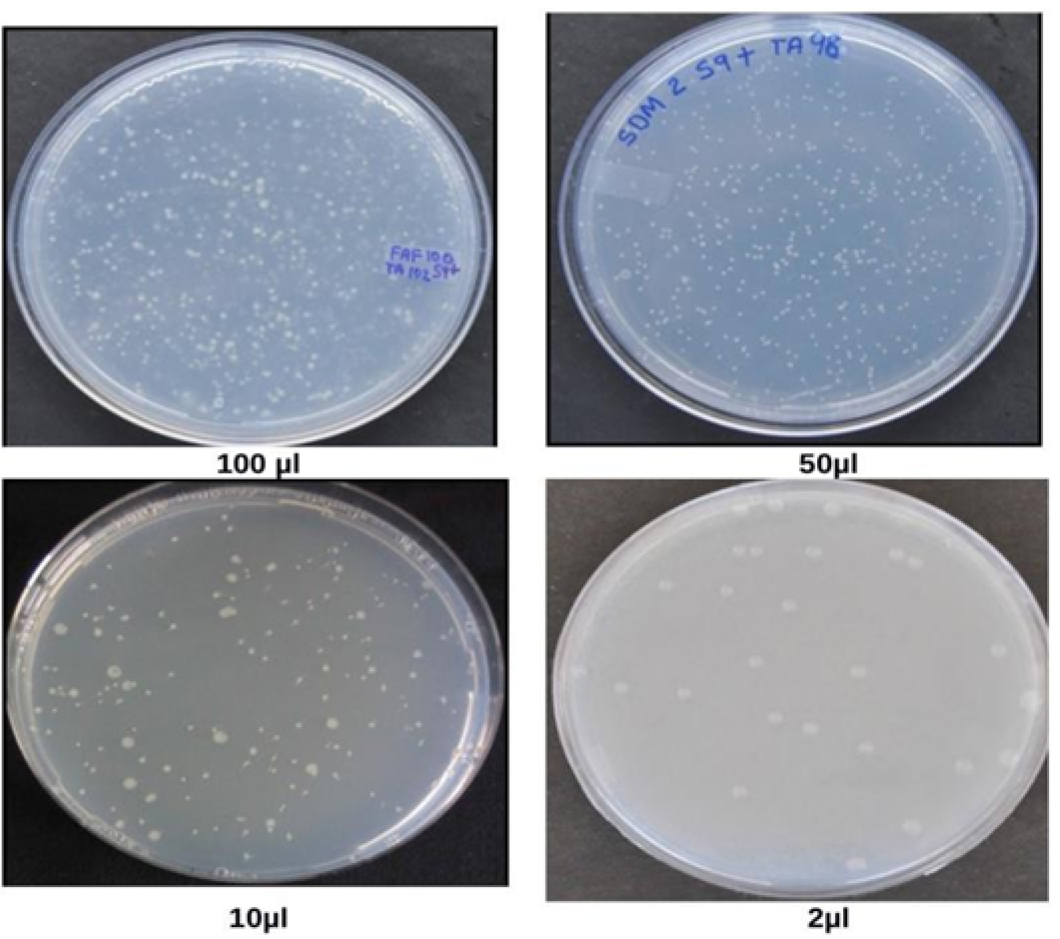
Figure 2. Spontaneous revertants colonies obtained after addition of waste water from health center in Salmonella mutagenicity assay at different concentrations, viz. 2 µl, 10 µl, 50 µl, 100 µl (Vijay, 2014)
Materials and Reagents
- Materials
- Tips (1,000 µl, 200 µl, 10 µl) (Tarsons)
- Sterile Petri plates (HiMedia Laboratories, catalog number: PW001 )
- Erlenmeyer flask and beaker (SchottDuran,10 ml, 250 ml, 500 ml)
- Eppendorf tubes (Tarsons,1.5 ml, 2.0 ml)
- Metal loop holder (metal loop Ch-2, HiMedia Laboratories, catalog number: LA012 )
- L shaped spreader(HiMedia Laboratories, catalog number: PW1085 )
- Tips (1,000 µl, 200 µl, 10 µl) (Tarsons)
- Mutagens
- Sodium azide (HiMedia Laboratories, catalog number: GRM1038 )
- 4-Nitroquinoline N-oxide (Sigma-Aldrich, catalog number: N8141 )
- 2-Aminofluorene (Sigma-Aldrich, catalog number: A55500 )
- Benzo(a)pyrene (Sigma-Aldrich, catalog number: B1760 )
- Mitomycin C (Roche Diagnostics, catalog number: 10107409001 )
- 2,4,7-Trinitro-9-fluorenone (Accustandard, catalog number: R-033S )
- 4-Nitro-o-phenylenediamine (Sigma-Aldrich, catalog number: 108898 )
- Sodium azide (HiMedia Laboratories, catalog number: GRM1038 )
- Reagents
- Oxoid nutrient broth No. 2 (Sigma-Aldrich, catalog number: 70123 )
Note: This product has been discontinued. - 70% ethanol
- Magnesium sulphate heptahydrate (MgSO4·7H2O) (HiMedia Laboratories, catalog number: RM683 )
- Citric acid monohydrate (HiMedia Laboratories, catalog number: GRM1008 )
- Potassium phosphate, dibasic (K2HPO4) (anhydrous) (Merck, catalog number: 61788005001730 )
- Sodium ammonium phosphate tetrahydrate (NaNH4HPO4·4H2O) (Sigma-Aldrich, catalog number: S9506 )
- D-biotin (HiMedia Laboratories, catalog number: TC096 )
- L-histidine (HiMedia Laboratories, catalog number: TC076 )
- Hydrochloric acid (HCI) (HiMedia Laboratories, catalog number: AS003 )
- Potassium chloride (KCl) (Merck, catalog number: 61753305001730 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (HiMedia Laboratories, catalog number: MB040 )
- Sodium dihydrogen phosphate monohydrate (NaH2PO4·H2O) (Merck, catalog number: 1063700050 )
- Disodium hydrogen phosphate (Na2HPO4) (HiMedia Laboratories, catalog number: TC051 )
- NADP (sodium salt) (HiMedia Laboratories, catalog number: RM392 )
- D-glucose-6-phosphate (monosodium salt) (Sigma-Aldrich, catalog number: G7879 )
- Ampicillin trihydrate (Sigma-Aldrich, catalog number: A6140 )
- Sodium hydroxide (NaOH) (Merck, catalog number: 106462 )
- Crystal violet (Sigma-Aldrich, catalog number: C6158 )
- Agar-Agar (Himedia Laboratories, catalog number: RM026 )
- Nutrient broth (HiMedia Laboratories, catalog number: M002 )
- Tetracycline (Sigma-Aldrich, catalog number: 87128 )
- Dimethylsulfoxide (HiMedia Laboratories, catalog number: TC185 )
- Vogel-Bonner medium E (50x) (see Recipes)
- 0.5 mM histidine/biotin solution (see Recipes)
- Salt solution (1.65 M KCl + 0.4 M MgCl2) (see Recipes)
- 0.2 M sodium phosphate buffer, pH 7.4 (see Recipes)
- 1 M Nicotinamide Adenine Dinucleotide Phosphate (NADP) solution (see Recipes)
- 1 M glucose-6-phosphate (see Recipes)
- Ampicillin solution (4 mg/ml) (see Recipes)
- Crystal violet solution (0.1%) (see Recipes)
- Minimal glucose plates (see Recipes)
- Histidine/Biotin plates (see Recipes)
- Ampicillin and tetracycline* plates (see Recipes)
- Nutrient agar plates (see Recipes)
- S9 mix (Rat Liver Microsomal Enzymes + Cofactors) (see Recipes)
- Sodium azide (see Recipes)
- Mitomycin (see Recipes)
- 2-Anthramine (see Recipes)
- Oxoid nutrient broth No. 2 (Sigma-Aldrich, catalog number: 70123 )
Equipment
- Orbital shaking incubator (Remi, model: RIS-24(BL) )
- Laminar Flow hood (Bio safety cabinet) (Deepak Meditech Pvt Ltd., Steri clean)
- Pipettes (Eppendorf, model: Research® plus, catalog number: 3120000062 , 1,000 μl; catalog number: 3120000046 , 200 μl; catalog number: 3120000020 , 10 μl)
- Vortex mixer (Labnet International, catalog number: S0100 )
- Hot water bath (Daiki Sciences, catalog number: KBLee2001 )
- Autoclave (TSC)
- Automatic Colony counter (Sonar)
- Refrigerator centrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: Heraeus Biofuge Primo R )
- pH meter (Labindia Analytical Instruments, model: PICO pH Meter , catalog number: PC13330101)
- Tissue tearor (Bio Spec Products, catalog number: 985370-04 )
Procedure
- Before performing the experiment, inoculate a single fresh colony of standard strains of S. typhimurium TA 98, 100 and 102, in oxoid nutrient broth-2 and incubate for 10-12 h at 37 °C in an incubator shaker at 120 rpm to ensure sufficient aeration for 1 x 109 bacterial cells. Each strain of S. typhimurium is grown separately in Erlenmeyer flasks (10 ml).
- Prepare fresh mutagen for each experiment (see Recipes).
Negative control: Autoclaved distilled water
Positive controls for TA 98, TA 100 and TA 102 without S9 metabolic activation (S9 mix): sodium azide (1 μg/ml) 2-nitrofluorine (1 μg/ml) and mitomycin (0.125 μg/ml)
For TA 98, TA 100 and TA 102 with S9 metabolic activation (S9 mix): 2-Anthramine (2 μg/ml) - Preparation of minimal glucose agar (MGA) plates: Mix the medium of minimal glucose agar plates (Recipe 9) and pour 25 ml into each Petri dish. Prepare the plates freshly before use.
- Label all minimal glucose agar plates and Eppendorf tubes prior to experiment.
- To the 2 ml sterile Eppendorf tubes, add the following each:
- 0.1 ml fresh culture of Salmonella strains
- 0.2 ml of His/Bio solution
- 0.5 ml sodium phosphate buffer (absence of S9 mix) or 0.5 ml S9 (presence of S9 mix)
- 0.1 ml of test sample or 0.1 ml of positive or negative control
- Make up to 1 ml with autoclaved distilled water.
- 0.1 ml fresh culture of Salmonella strains
- Mix the contents of Eppendorf tubes and pour onto Petri plates and spread using L-shaped spreader on the surface of MGA plates. Cover the Petri plates with sterile aluminum foil to protect the testing sample from photo reactive substances.
- After incubation of 48 h at 37 °C, spontaneous revertants colonies appear and are clearly visible with unaided eyes. All plates are run in triplicates.
- Revertants form a uniform lawn of auxotrophic bacteria on the surface the background of medium.
Data analysis
Non-statistical analysis
The most widely used method for non-statistical analysis of result in Ames test is ‘two-fold rule’ described by Mortelmans and Zeiger (2000) and Morino-Caniello and Piegorsch (1996). On the basis that the increase in the number of revertant colonies, the concentration of the tested sample goes up (dose-dependent manner), mutagenicity ratio (MR) is calculated first by counting the number of revertant colonies per plate and then calculating the MR as described by Maron and Ames (1983) using the formula below (see Sample data below for results):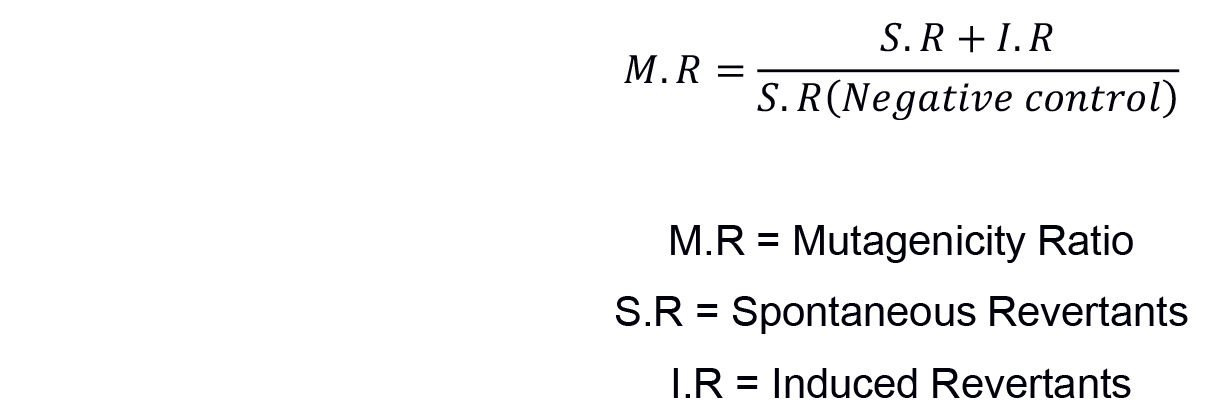
Sample data
Medical liquid waste was collected from different health care premises of Jaipur city. Salmonella mutagenicity test was performed on all the samples in their crude natural state using the plate incorporation procedure described by Maron and Ames, 1983. The results of Salmonella mutagenicity assay was analyzed through Mutagenicity Ratio method and shown in Table 4.
Table 4. Mutagenicity ratios of S. typhimurium strains TA98, TA100 and TA102 treated with waste water from different health premises (Vijay, 2014)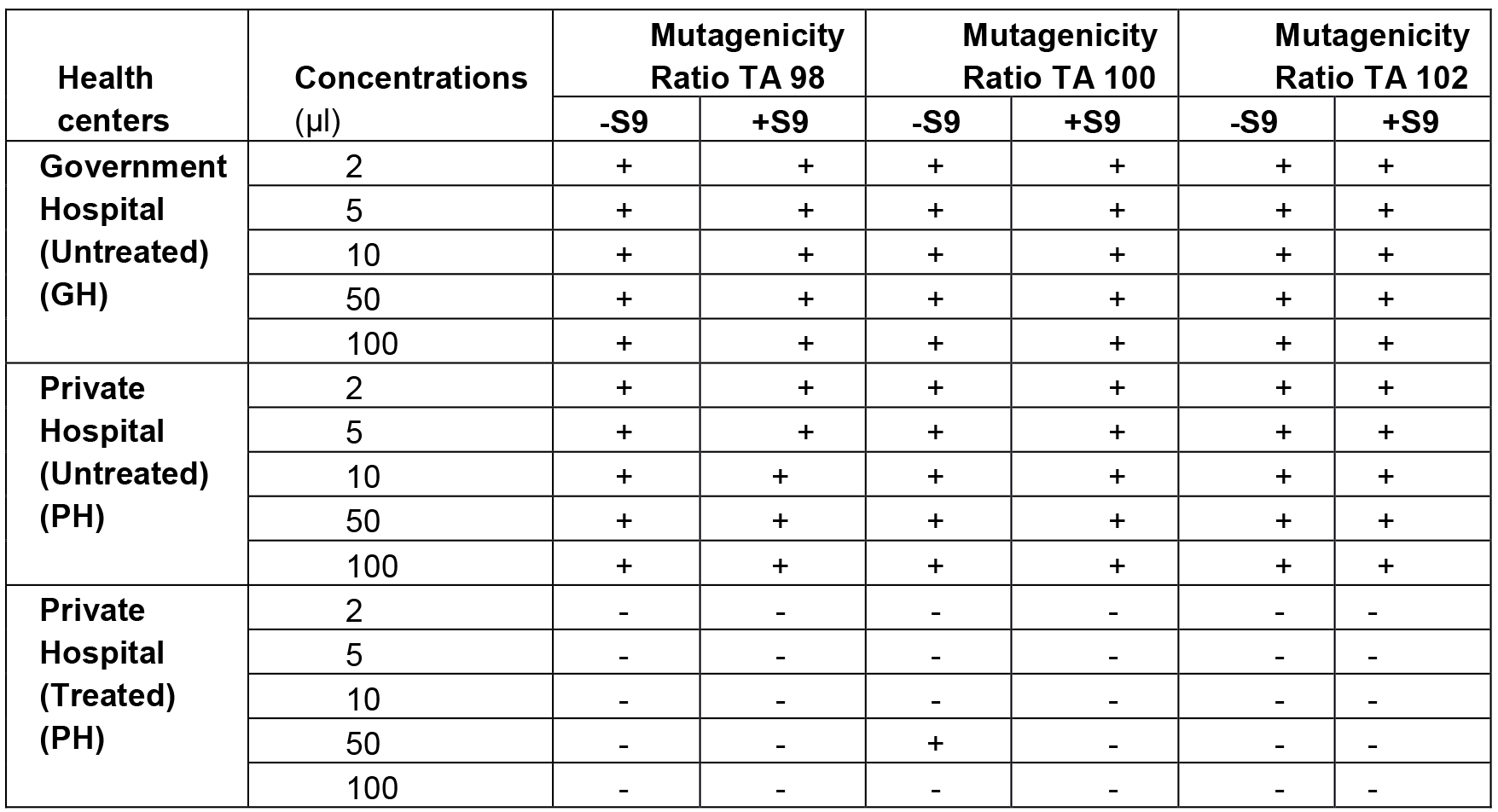
+Mutagenicity Ratio > 2.0 imply mutagenic, -Ratio < 2.0 imply non-mutagenic
Conclusion
The Ames test is a widely accepted bacterial assay to detect the mutagenicity in pathogenic bacteria. In this protocol, although we have shown the step wise methodology to perform Ames assay applicable for three strains, this method can be used for studying all compounds to infer mutagenicity. Whereas the Ames assay experiments involve sterile measures, care must be taken in ensuring the sample/plasmid is not contaminated. The improved methods to detect the genotoxicity of compounds help us troubleshoot methods for studying the compounds tested in clinical trials.
Notes
Sterilization (safety considerations while working with Salmonella)
- As S. typhimurium is a pathogenic bacterium, it is prudent to use precautionary measures every time and apply standard biosafety guidelines such as using plugged pipettes, proper sterilization by 70% ethanol and autoclaving all contaminated material.
- Handling of chemicals and strains should be done in biosafety cabinet. Before and after the use, cabinet must be sterilized using 70% ethanol and exposed to 15 min UV.
- Care must be taken to protect from chemical exposure by wearing gowns, eye glasses and gloves.
- Before discarding, all contaminated material (e.g., test tubes, pipettes and pipette tips, gowns and gloves) should be properly autoclaved.
Limitations
Ames assay consists of Salmonella typhimurium strains and so it is not a perfect model for human. Mice liver S9 hepatic fraction is used to minimize the mammalian metabolic activations formed in the hepatic system so that the mutagenicity of metabolites can be assessed. There are several differences between human and mice metabolism which can affect the mutagenicity of testing substances. Major disadvantages of fluctuation test is slower and slightly more laborious than Ames protocol. The test is primarily used for testing aqueous samples containing low levels of mutagen and therefore, this test is well adapted for evaluating the mutagenicity of wastewater samples.
Recipes
- Vogel-Bonner medium E (50x)
For Minimal agar (Recipe 9)Ingredients Per 500 ml Warm distilled H2O (45 °C) 335 ml Magnesium sulfate (MgSO4·7H2O) 5 g Citric acid monohydrate 50 g Potassium phosphate, dibasic (anhydrous) (K2HPO4) 250 g Sodium ammonium phosphate (NaNH4HPO4·4H2O) 87.5 g - Salts are added to the warm water in a flask. Place the flask on a hot plate
- After each salt dissolves entirely, transfer the solution into glass bottles and autoclave for 20 min at 121 °C
- When the solution gets cool, cap the bottle tightly
- Store the solution at 4 °C
- Salts are added to the warm water in a flask. Place the flask on a hot plate
- 0.5 mM histidine/biotin solution
For mutagenic bioassay
Dissolve the biotin in hot distilled water. The solution is autoclaved for 20 min, at 121 °C and then stored at 4 °CIngredients Per 125 ml D-Biotin (F.W. 247.3) 15.45 mg L-Histidine·HCl (F.W. 191.7) 12.0 mg Distilled H2O 125 ml - Salt solution (1.65 M KCl + 0.4 M MgCl2)
For S9 hepatic fraction
All the components are dissolved in water. The solution is autoclaved for 20 min, at 121 °C and then stored at 4 °CIngredients Per 250 ml Potassium chloride (KCl) 30.75 g Magnesium chloride (MgCl2·6H2O) 20.35 g Distilled H2O to final concentration of 250 ml - 0.2 M sodium phosphate buffer, pH 7.4
For S9 hepatic fraction
Adjust pH to 7.4. Sterilize the buffer by autoclaving for 20 min at 121 °CIngredients Per 250 ml 0.2 M sodium dihydrogen phosphate (NaH2PO4·H2O) 30 ml (6.9 g/250 ml) 0.2 M disodium hydrogen phosphate (Na2HPO4) 220 ml (7.1 g/250 ml) - 1 M nicotinamide adenine dinucleotide phosphate (NADP) solution
For S9 hepatic fraction
NADP is dissolved in the distilled water and mixed by vortexing. Tubes are placed in an ice bath. The solution can be used for up to six monthsIngredients Per 2.5 ml NADP 191.5 mg Sterile distilled H2O 2.5 ml - 1 M glucose-6-phosphate
For S9 hepatic fraction
Glucose-6-phosphate is dissolved in the 5 ml distilled water and mixed by vortexing. Tubes are placed in an ice bath. The solution can be used for up to six monthsIngredients Per 5 ml Glucose-6-phosphate (G-6-P) 1.41 g Sterile distilled H2O 5 ml - Ampicillin solution (4 mg/ml)
Used in tests of ampicillin resistance
Master plates for R-factor strains
Ampicillin trihydrate is dissolved in the 50 ml of NaOH (0.02 N) and mixed by vortexing. Tubes are placed in an ice bathIngredients Per 500 ml Ampicillin trihydrate 0.4 g Sodium hydroxide (0.02 N) 50 ml - Crystal violet solution (0.1%)
Used in tests for crystal violet sensitivity (to confirm rfa mutation)Ingredients Per 500 ml Crystal violet 0.05 g Distilled H2O 50 ml - Minimal glucose plates
Used in Mutagenic bioassay
Add agar in 465 ml of distilled water and autoclave for 20 min, at 121 °C. After cooling, add the salts and glucose gentlyIngredients Per 500 ml Agar 7.5 g Distilled H2O 465 ml 50x VB salts (Recipe 1) 10 ml 40% glucose 25 ml - Histidine/Biotin plates (Master plates for non R-factor strains)
Used in tests for histidine requirement
Dissolve agar in the given concentration in distilled water. Autoclave each solution separately for 20 min. After cooling of solution, add each salt gentlyIngredients Per 500 ml Agar 7.5 g Distilled H2O 457 ml 50x VB salts 10 ml 40% glucose 25 ml Sterile histidine (2 g per 400 ml H2O) 5 ml Sterile 0.5 mM biotin 3 ml - Ampicillin and tetracycline* plates
Master plates for the cultivation of strains containing the plasmids pKM101 and pAQ1*
Dissolve agar in the given concentration in distilled water. Autoclave each solution separately for 20 min. After cooling of solution, add each salt gentlyIngredients Per 500 ml Agar 7.5 g Distilled H2O 405 ml 50x VB salts 10 ml 40% glucose 25 ml Sterile histidine (2 g per 400 ml H2O) 5 ml Sterile 0.5 mM biotin 3 ml Sterile ampicillin solution (8 mg/ml 0.02 N NaOH) 1.58 ml *Sterile tetracycline solution (8 mg/ml 0.02 N HCl) 0.125 ml
*Note: TA 102 is resistant to tetracycline. The shelf life of the plates is two weeks at 4 °C. - Nutrient agar plates
Used in tests for genotypes [Crystal violet sensitivity (rfa) and UV sensitivity (AuvrB)] and viability of bacteria
Dissolve agar in the given concentration in distilled water. Autoclave separately for 20 min. Pour the cooled solution into the Petri platesIngredients Per 500 ml Nutrient agar 7.5 g Distilled H2O 500 ml - S9 mix (Rat Liver Microsomal Enzymes + Cofactors)
Note: Add each ingredient in the reverse order listed above (First water, and then phosphate buffer…). Avoid refreezing the S9 mix.Ingredients Standard S9 mix Per 25 ml Mice liver 1.0 ml (2%) MgCl2-KCl salts 0.5 ml 1 M glucose-6-phosphate 0.125 ml 0.1 M NADP 1.0 ml 0.2 M phosphate buffer, pH 7.4 12.5 ml Sterile distilled H2O 9.86 ml - Sodium azide
Used in Mutagenicity assay
Working concentrations are prepared by taking 1, 2, 4 µl of 10 mg/mlIngredients Per ml Sodium azide 10 µg Autoclave distilled H2O 990 µl (to make a total volume of 1 ml) - 2-Nitrofluorine
Used in Mutagenicity assay
Working concentrations are prepared by taking 1, 2, 4 µl of 10 mg/mlIngredients Per ml 2-Nitrofluroine 10 µg Autoclave distilled H2O 990 µl (to make a total volume of 1 ml) - Mitomycin
Used in Mutagenicity assay
Working concentrations are prepared by taking 1, 2, 4 µl of 10 mg/mlIngredients Per ml Mitomycin 10 µg Autoclave distilled H2O 990 µl (to make a total volume of 1 ml) - 2-Anthramine
Used in Mutagenicity assay
Working concentrations are prepared by taking 1, 2, 4 µl of 10 mg/mlIngredients Per ml 2-Anthramine 10 µg Autoclave distilled H2O 990 µl (to make a total volume of 1 ml)
Acknowledgments
Urvashi Vijay would like to thank the Department of Zoology, The IIS University, Jaipur where the work was carried out. The financial help received by IISU Fellowship 2012/9389 to Urvashi Vijay is gratefully acknowledged.
Conflict of interests: None declared.
Authors contributions: Urvashi Vijay, Sonal Gupta, Priyanka Mathur carried out the protocol and the methods under the guidance of Pradeep Bhatnagar. Prashanth Suravajhala re-reviewed the works and proofread the manuscript before all authors approving it.
References
- Ames, B. N. (1971). The detection of chemical mutagens with enteric bacteria. In: Hollaender, A. (Ed.). Chemical Mutagens, Principles and Methods for Their Detection vol. 1. Plenum pp: 851-863.
- Ames, B. N., Durston, W. E., Yamasaki, E. and Lee, F. D. (1973a). Carcinogens are mutagens: a simple test system combining liver homogenates for activation and bacteria for detection. Proc Natl Acad Sci U S A 70(8): 2281-2285.
- Ames, B. N., Lee, F. D. and Durston, W. E. (1973b). An improved bacterial test system for the detection and classification of mutagens and carcinogens. Proc Natl Acad Sci U S A 70(3): 782-786.
- Ames, B. N., McCann, J. and Yamasaki, E. (1975a). Methods for detecting carcinogens and mutagens with the Salmonella/mammalian-microsome mutagenicity test. Mutat Res 31(6): 347-364.
- Ames, B. N., Yamasaki, E., Mc Cann, J. (1975b). Detection of carcinogens in the Salmonella/microsome test. Assay of 300 chemicals. Proc Natl Acad Sci U S A 72: 5135-5139.
- Barnes, W., Tuley, E and Eisenstadt, E. (1982). Base sequence analysis of His+ revertants of the hisG46 missense mutations in Salmonella typhimurium. Enviorn Mutagen 4:297.
- Bernstein, L. S., Kaldor, J., McCann, J. and Pike, M. C. (1982). An empirical approach to the statistical analysis of mutagenesis data from the Salmonella test. Mutat Res 97: 267-281.
- Brusick, D. J., Simmon, V. F., Rosenkranz, H. S., Ray, V. A. and Stafford, R. S. (1980). An evaluation of the Escherichia coli WP2 and WP2 uvrA reverse mutation assay. Mutat Res 76(2): 169-190.
- Czyz, A., Szpilewska, H., Dutkiewicz, R., Kowalska, W. A., Biniewska-Godlewska, A., and Wegrzyn, G. (2002). Comparison of the Ames test and a newly developed assay for detection of mutagenic pollution of marine environments. Mutat Res 519: 67-74.
- DeFlora, S., Zanacchi, P., Camoirano, S., Bennicelli, S., Badolati, G. S. (1984). Genotoxic activity and potency of 135 compounds in the Ames reversion test and in a bacterial DNA-repair test. Mutat Res 133: 161-198.
- Fluckiger-Isler. S., Baumeister, M., Braun, K., Gervais, V., Hasler-Nguyen, N., Reimann, R., Van Gompel, J., Wunderlich, H. G. and Engelhardt, G. (2004). Assessment of the performance of the Ames II assay: a collaborative study with 19 coded compounds. Mutat Res 558: 181-197.
- Frantz, C. N and Malling, H. V. (1975). The quantitive microsomal mutagenesis assay method. Mutat Res 31: 365-380.
- Garner, R. C., Miller, E. C. and Miller, J. A. (1972). Liver microsomal metabolism of aflatoxin B 1 to a reactive derivative toxic to Salmonella typhimurium TA 1530. Cancer Res 32(10): 2058-2066.
- Gupta, P., Mathur, N., Bhatnagar, P., Nagar, P. and Srivastava, S. (2009). Genotoxicity evaluation of hospital wastewaters. Ecotoxicol Environ Saf 72(7): 1925-1932.
- Hayes, A. W. (1982). Principles and methods of toxicology. Raven Press pp: 238-241.
- Henderson, L., Albertini, S. and Aardema, M. (2000). Thresholds in genotoxicity responses. Mutat Res 464(1): 123-128.
- Levin, D. E., Yamasaki, E. and Ames, B. N. (1982). A new Salmonella tester strain, TA97, for the detection of frameshift mutagens. A run of cytosines as a mutational hot-spot. Mutat Res 94(2): 315-330.
- Maron, D. and Ames, B. N. (1983). Revised methods for the Salmonella mutagenicity test. Mutat Res 113: 173-215.
- Maron, D., Katzenellenbogen, J. and Ames, B. N. (1981). Compatibility of organic solvents with the Salmonella/microsome test. Mutat Res 88: 343-350.
- Mathur, N., Bhatnagar, P. and Bakre, P. (2005). Assessing mutagenicity of textile dyes from Pali (Rajasthan) using Ames Bioassay. Appl Ecol Env Res 4: 111-118.
- Morino-Caniello, N. F. and Piegorsch, W. W. (1996). The Ames test: the two-fold rule revisited. Mutat Res 369: 23-31.
- Mortelmans, K. and Stocker, B. A. D. (1979). Segregation of the mutator property of plasmid R46 from its ultraviolet-protection properties. Mol Gen Genet 167: 317-327.
- Mortelmans, K. and Zeiger, E. (2000). The Ames Salmonella/microsome mutagenicity assay. Mutat Res 455: 29-60.
- Prival, M. J., Bell, S. J., Mitchell, V. D., Peiperl, M. D. and Vaughan, V. L. (1984). Mutagenicity of benzidine and benzidine-congener dyes and selected monoazo dyes in a modified Salmonella assay. Mutat Res 136(1): 33-47.
- Shanabruch, W. G. and Walker, G. C. (1980). Localization of the plasmid (pKMIOl) gene(s) involved in recA+/ex4+-dependent mutagenesis. Mol Gen Genet 179: 289-297.
- Simmon, V. F., Kauhanen, K. and Tardiff, R. G. (1977). Mutagenic activitiesof chemicals identified in drinking water. In: Progress in Genetic Toxicology. 249-258.
- Venitt, S. and Bosworth, D. (1983). The development of anaerobic methods for bacterial mutation assays: aerobic and anaerobic fluctuations tests of human faecal extracts and reference mutagens. Carcinogenesis 4: 339-345.
- Vijay, U., Bhatnagar, P. and Mathur, P. (2014). Mutagenicity Evaluation of Health Center wastewater with the mutant TA 100 and TA 102 strains of Salmonella typhimurium. Inventi Impact: Pharm Biotech & Microbio 4: 195-198.
- Vijay, U. (2014). Physico-chemical characterization and toxicological evaluation of liquid effluents generated by health care establishments of Jaipur. The IIS University. Thesis
- Walker, G. C. and Dobson, P. P. (1979). Mutagenesis and repair deficiencies of Escherichia coli umuC mutants are suppressed by the plasmid pKM101. Mol Gen Genet 172: 17-24.
- Wilcox, P., Naidoo, A., Wedd, D. J. and Gatehouse, D. G. (1990). Comparison of Salmonella typhimurium TA102 with Escherichia coli WP2 tester strains. Mutagenesis 5(3): 285-291.
- Zeiger, E. (1985). The Salmonella mutagenicity assay for identification of presumptive carcinogens. In: Milman, H. A. and Weisburger, E. K. (Eds). Handbook of Carcinogen Testing. Noyes Publishers pp: 83-99.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Vijay, U., Gupta, S., Mathur, P., Suravajhala, P. and Bhatnagar, P. (2018). Microbial Mutagenicity Assay: Ames Test. Bio-protocol 8(6): e2763. DOI: 10.21769/BioProtoc.2763.
Category
Microbiology > Microbial genetics > Mutagenesis
Molecular Biology > DNA > DNA damage and repair
Cell Biology > Cell isolation and culture > Cell growth
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










