- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analysis of Chromosome Condensation/Decondensation During Mitosis by EdU Incorporation in Nigella damascena L. Seedling Roots
Published: Vol 8, Iss 3, Feb 5, 2018 DOI: 10.21769/BioProtoc.2726 Views: 6898
Reviewed by: Amey RedkarSmita NairIsabelle Colas

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Visualization of Actin Organization and Quantification in Fixed Arabidopsis Pollen Grains and Tubes
Xiaolu Qu [...] Shanjin Huang
Jan 5, 2020 5339 Views
Acidified Blue Ink-staining Procedure for the Observation of Fungal Structures Inside Roots of Two Disparate Plant Lineages
Jill Kowal [...] Sophie Lane
Oct 20, 2020 5026 Views
Live Leaf-Section Imaging for Visualizing Intracellular Chloroplast Movement and Analyzing Cell–Cell Interactions
Yuta Kato [...] Mitsutaka Taniguchi
Aug 5, 2025 2301 Views
Abstract
To investigate the chromosome dynamics during mitosis, it is convenient to mark the discrete chromosome foci and then analyze their spatial rearrangements during prophase condensation and telophase decondensation. To label the chromosome regions in plant chromosomes, we incorporated the synthetic nucleotide, 5-ethynyl-2’-deoxyuridine (EdU), which can be detected by click-chemistry, into chromatin during replication. Here, we described a protocol of a method based on the application of semi-thin sections of Nigella damascena L. roots embedded in LR White acrylic resin. The thickness of semi-thin (100-250 nm) sections is significantly lower than that of optical sections even if a confocal microscope was used. This approach may also be suitable for work with any tissue fragments or large cells (oocytes, cells with polytene chromosomes, etc.).
Keywords: PlantBackground
Most data concerning chromosome organization have been acquired from studies of a small number of model organisms, the majority of which are mammals. In plants with large genomes, the chromosomes are significantly larger than the animal chromosomes that have been studied to date. To investigate the chromosome dynamics during mitosis, it is necessary to mark the discrete chromosome foci and then analyze their spatial rearrangements during prophase condensation and telophase decondensation. To label the chromosome regions, we incorporated the synthetic nucleotide, 5-ethynyl-2’-deoxyuridine (EdU) (Kuznetsova et al., 2017). Detection of EdU is based on a click-reaction, which is a copper catalyzed reaction between an azide and an alkyne. The EdU contains the alkyne which can react with the azide-containing detection reagent.
The most suitable distribution of labeled regions (i.e., separated labeled dots) was seen in cells which incorporated EdU during late S-phase. The brief pulse labeled all S-phase cells, and the initial appearance of EdU-labeled mitotic figures thus denoted the time needed for cells labeled in late S-phase to traverse into mitosis.
Root apical meristem does not allow for the acquisition of high-resolution images because of the out-of-focus fluorescence. Here, we described a protocol of a method based on the application of semi-thin (100-250 nm) sections of roots embedded in LR White acrylic resin. LR White is a polyhydroxy-aromatic acrylic resin with low toxicity and ultra-low viscosity. The polymerized resin is hydrophilic (sections freely permeable to aqueous solutions). The thickness of semi-thin sections is significantly lower than that of optical sections even if a confocal microscope was used. This approach may also be suitable for work with any tissue fragments or large cells (e.g., cells with polytene chromosomes).
Materials and Reagents
- 30 mm and 90 mm Petri dishes (Greiner Bio One International)
- Filter paper
- Single edge blades (Ted Pella, catalog number: 121-3 )
- 1.5 microtubes (SSIbio, catalog number: 1260-00 )
- Snap-fit Gelatin Capsules, Size 2 (Ted Pella, catalog number: 130-19 )
- Cover slips, No. 1 (Fisher Scientific, catalog number: 12-548A )
- Microscope slides (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: AA00000112E00MNT10 )
- Parafilm
- Cotton wool
- Foil
- Safety gloves
- Nigella damascena L. seeds
- Click-iT EdU Alexa 555 Imaging Kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: C10338 )
- Thymidine (Sigma-Aldrich, catalog number: T9250 )
- Phosphate buffered saline (PBS), pH 7.2 (10x) (Thermo Fisher Scientific, GibcoTM, catalog number: 70013016 )
- Ethanol
- 4’,6-Diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 62248 )
- Tris base
- Concentrated HCl
- Paraformaldehyde (Sigma-Aldrich, catalog number: P6148 )
- LR White embedding kit (Sigma-Aldrich, catalog number: 62662 )
Note: This product has been discontinued. - Formvar 1595E (Serva, catalog number: 21740 )
- 1,2-Dichloroethane anhydrous (Sigma-Aldrich, catalog number: 284505 )
- Mowiol 4-88 (Sigma-Aldrich, catalog number: 81381 )
- Glycerol (MP Biomedicals, catalog number: 04800687 )
- 1,4-Diazabicyclo-[2.2.2]-octane (Sigma-Aldrich, catalog number: D2522 )
Note: This product has been discontinued. - Deionized H2O
- 1 M Tris-HCl (pH 8.5) (see Recipe 1)
- Paraformaldehyde (see Recipe 2)
- LR White acrylic resin (see Recipe 3)
- Formvar coated cover slips (see Recipe 4)
- Mowion mounting medium (see Recipe 5)
Equipment
- Perfect Loop (Ted Pella, catalog number: 13064 )
- Orbital Shaker OS-20 (Biosan, model: OS-20 , catalog number: BS-010108-AAG)
- Laboratory incubator TC1/20 (SKTB, model: TC-1/20 , catalog number: 1003)
- Chemical fume hood
- Ultratome LKB III
- Fluorescent microscope Axiovision 200M (Carl Zeiss, model: Axiovision 200M ) equipped with the ORCAII-ERG2 camera (Hamamatsu).
Note: For deconvolution, AxioVision 3.1 software (Carl Zeiss) was used. - pH meter
- Magnetic stirrer with hot plate MSH-300 (Biosan, model: MSH-300, catalog number: BS-010302-OAA )
- Glass or diamond knife
- Micro-centrifuge MiniSpin (Eppendorf, model: MiniSpin® plus , catalog number: 5452000018)
Procedure
- Germinate the seeds of Nigella damascena L. in 90 mm Petri dishes covered with damp filter paper at 25 °С in the dark. It takes about a week for roots to grow into 10 mm.
- For chromatin labeling, use 10 mm long roots. Incubate the germinated seeds in distilled water with 50 μM EdU (EdU is a part of Click-iT EdU Alexa 555 Imaging Kit, prepare the stock solution according to manufacturer’s recommendations) for 30 min. Incubate in 90 mm Petri with 10 ml of EdU solution on an orbital shaker (50 rpm) mounted inside an incubator at 25 °С.
- Incubate seeds in 200 µM thymidine for 30 min, and then grow in distilled water during different time period that allowed to label different chromosome regions. In case of Nigella damascena L., 3.5 h incubation is sufficient to find the first labeled mitosis (Kuznetsova et al., 2017). Incubate in 90 mm Petri with 10 ml of thymidine solution or distilled water on an orbital shaker (50 rpm) mounted inside an incubator at 25 °С.
- Excise root tips that are 1.0 mm long from 10 mm long roots using a single edge blade, transfer to 1.5 ml microtubes and fix in 2% paraformaldehyde in 0.5x PBS for 1.5 h at room temperature (for additional fixation protocol see Note 1).
Note: Use a chemical fume hood while handing formaldehyde-containing chemicals. - After fixation, wash the root tips in 0.5x PBS, dehydrate and infiltrate in LR White acrylic resin using the following steps:
- Wash roots with 0.5x PBS 3 times for 5 min each.
- Wash roots with 50% ethanol for 30 min.
- Dehydrate specimens in 70% ethanol 2 times for 30 min each (it is possible to store specimens in 70% ethanol at 4 °С).
- Infiltrate with LR White and 70% ethanol (2:1) mixture (1 ml) for 1 h.
For LR white/ethanol mixture preparation, slowly add one part of 70% ethanol (drop by drop) to two parts of LR White, and rotate the tube gently (otherwise the mixture will become milky). - Incubate in LR White (1 ml) for 1 h.
- Incubate in LR White (1 ml) overnight at 4 °С.
- Incubate in LR White (1 ml) for 1 h.
- Wash roots with 0.5x PBS 3 times for 5 min each.
- It is important to limit the contact of oxygen with the resin while polymerization occurs. The most convenient way of achieving this is to use gelatin capsules. Place several (3-4) roots in the bottom of the capsule and fill up with LR White to the brim and slide the other half of the capsule on (Figure 1).
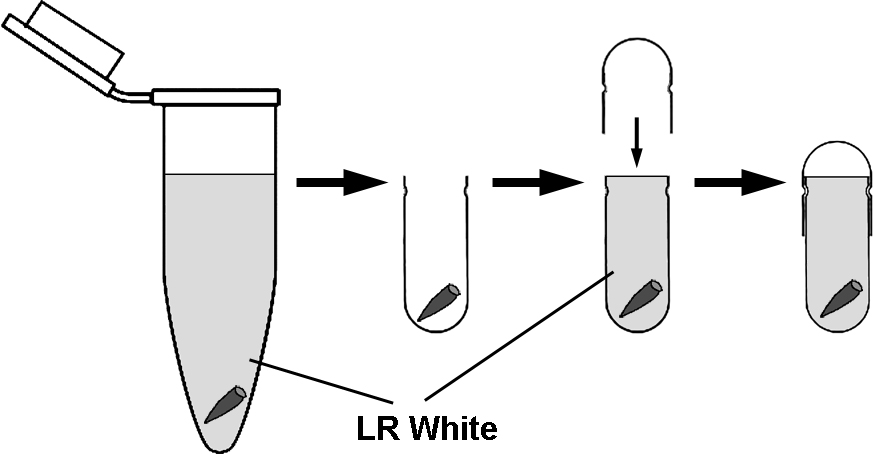
Figure 1. The transfer of roots from microtubes to gelatin capsules for polymerization. For removal of roots from microtubes and transfer to capsules, it is convenient to use straight tip tweezers and do not discard LR White form the microtubes. Fill up the gelatin capsules with LR White to the brim. - Polymerize at 55 °С for 24 h.
- Prepare semi-thin sections (100-250 nm) using an Ultratome using either glass knife or diamond knife for sectioning of histology specimens.
- Mount semi-thin sections on Formvar-coated cover slips (see Recipes) using a Perfect Loop.
- Dry cover slips with semi-thin sections at 37 °С for 2-3 h for better attachment of section.
- Detect EdU using a Click-iT EdU Alexa 555 Imaging Kit according to manufacturer’s protocol (see Note 2). The staining is carried out in the humid chamber in the dark. The humid chamber is a 90 mm non-vented Petri dish, whose bottom is covered with Parafilm. A piece of moist cotton wool is placed near the dish wall. Drops of a dye solution are placed on the Parafilm, cover slips are placed on these drops (sections downwards) (Figure 2). The dishes are sealed with the Parafilm.

Figure 2. Humid chamber - Stain DNA with 1 μg/ml DAPI solution in 30 mm Petri dishes for 10 min, and after washing in PBS for 5 min, mount sections in Mowiol with the anti-bleaching agent, 1,4-diazabicyclo-[2.2.2]-octane (DABCO).
- Image stacks may be acquired using either wild-field fluorescent microscopy with 3D-deconvolution (Figure 3).
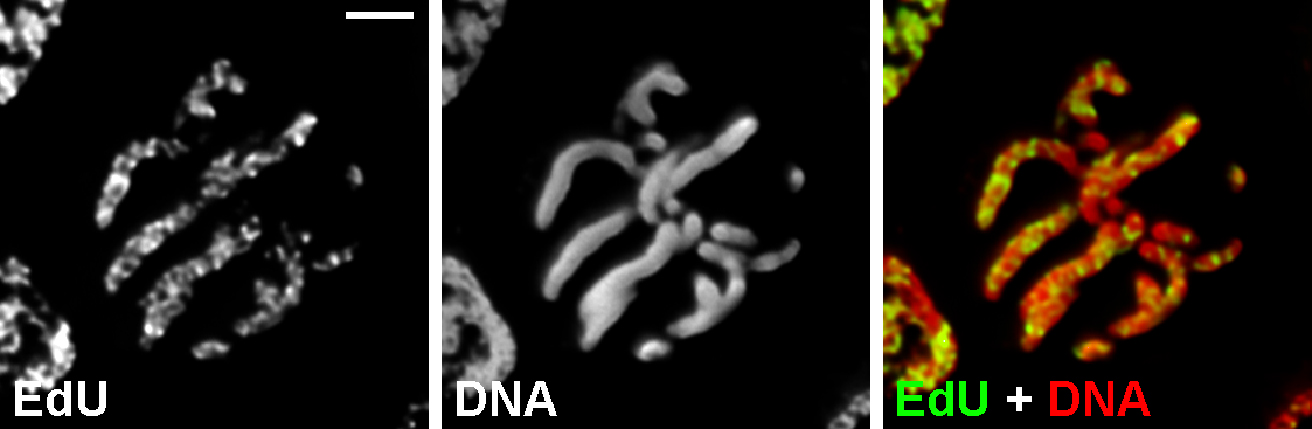
Figure 3. Chromosome labeling with EdU. Metaphase cell of Nigella damascena L. whose chromosomes included EdU during late S-phase. Wild-field fluorescent microscopy with 3D-deconvolution. Scale bar = 5 μm.
Notes
- It is possible to use glutaraldehyde for fixation (2.5% glutaraldehyde in 50 mM Sörensen buffer). But after such fixation, the pale autofluorescence is clearly visible. We cannot recommend this fixation for conventional light microscopy, but it is possible to use it for high-resolution microscopy and correlative light and electron microscopy.
- There are a lot of fluorophore azides from different manufactures (Molecular Probes, Lumiprobe, Sigma-Aldrich, etc.). The selection of fluorochrome for the staining depends on the microscopic system which is used for image acquisition. Our microscope has filter sets optimized for the detection of Alexa 488 and Alexa 555 (or different fluorochromes with similar spectral characteristics). We prefer to use Alexa 555 because of substantially brighter staining which we obtained with this fluorochrome.
Recipes
- 1 M Tris-HCl (pH 8.5)
- Dissolve Tris base in water (Tris base 121.14 g, deionized water ~800 ml)
- Using a pH meter, titrate the solution with concentrated HCl until the correct pH is reached
- Add deionized water to 1,000 ml
- Store at 4 °C
- Dissolve Tris base in water (Tris base 121.14 g, deionized water ~800 ml)
- Paraformaldehyde
- Mix 2.0 g of paraformaldehyde with 100 ml of 0.5x PBS in a glass that was tightly closed with foil
- Heat the solution on a magnetic stirrer with a hot plate until it becomes transparent (Figure 4). Do not let the solution boil. We prefer to heat with the maximal temperature with the permanent observation
- Then the solution will be transparent, it is necessary to remove the glass from the heater. The diluted formaldehyde may be aliquoted, frozen and stored at -20 °C. We never stored the aliquots for more than 1 month. It is important to thaw out each aliquot only one time

Figure 4. Paraformaldehyde dissolution using magnetic stirrer with hot plate - Mix 2.0 g of paraformaldehyde with 100 ml of 0.5x PBS in a glass that was tightly closed with foil
- LR White acrylic resin
- Add the benzoyl peroxide catalyst (1.998 g) to the LR white acrylic resin (100 ml) at room temperature (both reagents are the parts of LR White embedding kit)
- The catalyst will take a full 24 h at room temperature to dissolve completely. During this time, occasionally shaking the activated resin mixture is recommended. Once mixed with catalyst and fully dissolved, the resin must be stored at 2-8 °C to maintain its shelf life but not longer than 9 months
Note: Work under the chemical fume hood and in safety gloves. If you have direct contact with skin or eyes, wash the affected areas with plenty of water and soap. - Add the benzoyl peroxide catalyst (1.998 g) to the LR white acrylic resin (100 ml) at room temperature (both reagents are the parts of LR White embedding kit)
- Formvar coated cover slips
3 mg of Formvar is diluted in 10 ml of 1,2-dichloroethane. This solution can be stored in airtight glass in the dark. Cleaned cover slips are dipped in a solution of Formvar and left to dry vertically. Formvar coated cover slips should be prepared before the use - Mowion mounting medium
- Add 1.2 g of Mowiol 4-88 to 3 g of glycerol, and then add 3 ml of deionized H2O and leave for 2 h at room temperature
- Add 6 ml of 0.2 M Tris-HCl (pH 8.5) and heat to 50 °C with occasional mixing till the Mowiol will be dissolved. Add 1,4-diazabicyclo-[2,2,2]-octane (DABCO) to final concentration 2.5% to reduce fading
- Centrifuge the mounting medium at maximal speed in micro-centrifuge for 15 min, and transfer supernatant to the clean microtubes
- Aliquot in airtight microtubes can be stored at -20 °C
- Add 1.2 g of Mowiol 4-88 to 3 g of glycerol, and then add 3 ml of deionized H2O and leave for 2 h at room temperature
Acknowledgments
The work was supported by the Russian Science Foundation (project 14-15-00199). The protocol was adapted from previous work (Kuznetsova et al., 2017). The authors declare no conflicts of interest or competing interests that may impact the design and implementation of this protocol.
References
- Kuznetsova, M. A., Chaban, I. A. and Sheval, E. V. (2017). Visualization of chromosome condensation in plants with large chromosomes. BMC Plant Biol 17(1): 153.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sheval, E. V. (2018). Analysis of Chromosome Condensation/Decondensation During Mitosis by EdU Incorporation in Nigella damascena L. Seedling Roots. Bio-protocol 8(3): e2726. DOI: 10.21769/BioProtoc.2726.
Category
Cell Biology > Cell staining > Nucleic acid
Plant Science > Plant cell biology > Cell structure
Plant Science > Plant cell biology > Cell staining
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










