- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Large-scale Maize Seedling Infection with Exserohilum turcicum in the Greenhouse
Published: Vol 7, Iss 19, Oct 5, 2017 DOI: 10.21769/BioProtoc.2567 Views: 10320
Reviewed by: Zhibing LaiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
In Vitro Screening of Microbial Extracts Against the Oomycetes Phytophthora capsici and Pythium ultimum
Mónica Trigal Martínez [...] María Ángeles Vinuesa Navarro
Sep 20, 2025 1190 Views

Quantitative Estimation of Auxin, Siderophore, and Hydrogen Cyanide Production in Halo and Drought-Tolerant Bacterial Isolates for Cucumber Growth
Zeinab Fotoohiyan and Ali Salehi Sardoei
Oct 5, 2025 1267 Views

A Reliable In Planta Inoculation and Antifungal Screening Protocol for Rhizoctonia solani-Induced Sheath Blight in Rice
Alinaj Yasin [...] Palash Deb Nath
Nov 5, 2025 1448 Views
Abstract
Northern corn leaf blight (NCLB) is a serious foliar disease of maize (Zea mays) worldwide and breeding for resistance is of primary importance for maize crop protection. Phenotyping for NCLB resistance is well established in the field, but such experiments depend on suitable environmental conditions and are seasonal. Here we describe a greenhouse seedling approach that is suitable for testing thousands of seedling plants in a single experiment with a duration of 37 days. Three scoring methods were used to quantify the disease severity: the area under the disease progress curve (AUDPC), the primary diseased leaf area of the inoculated leaves at 16 days post inoculation (PrimDLA at 16 dpi) and the incubation period (IP) that was determined as days from inoculation to symptom appearance. By testing a diverse panel of maize genotypes, a high correlation between the three different methods was observed (81.9% to 94.1%), indicating that each of scoring methods can be applied for disease quantification. Thus, the seedling assay developed served as a relatively simple and high-throughput method for phenotyping NCLB disease resistance under greenhouse condition.
Keywords: Northern corn leaf blightBackground
Northern corn leaf blight (NCLB) is a ubiquitous foliar wilt disease that threatens maize production worldwide (Welz and Geiger, 2000). The disease is caused by the hemibiotrophic fungus Exserohilum turcicum (anamorph of Setosphaeria turcica), which favors a high-humidity and cool temperature environment. Under favorable conditions, fungal infection manifests itself as large and irregularly emerging lesions that destroy the entire foliage. Therefore, this disease decreases the active leaf area and the accumulation of photosynthesized products. Up to 50% grain yield loss was reported but the reduction largely depended on environmental parameters (e.g., temperature, humidity), phases of maize development and hybrid susceptibility (Ullstrup, 1970; Pataky et al., 1998).
Precision phenotyping for NCLB disease resistance is critical for the determination of host resistance against E. turcicum. Testing for disease resistance in the field is well established, e.g., by placing or distributing inoculums in the leaf whorl at the 4 to 6 leaf stage (or even older) plants (Dingerdissen et al., 1996; Lipps et al., 1997; Brown et al., 2001; Asea et al., 2009; Chung et al., 2010; Chung et al., 2011). Scoring for resistance can be conducted by determining the levels of susceptibility (1 to 9; 1, complete resistance, no symptoms; 9, 90-100% of leaf area infected), the primary diseased leaf area (PrimDLA) that was defined as the percentage of infected leaf area of the inoculated leaf, the diseased leaf area of the entire plant (DLA), the incubation period (IP) rated as the number of days post inoculation until first observing the wilting/lesion, the lesion number (LN) at 14 to 21 days post inoculation and finally the area under the disease progress curve (AUDPC). However, tests for resistance in the field are environmentally-dependent and time-consuming. Here we describe a simple greenhouse seedling assay by testing only the second leaf, thus being suitable for quantifying thousands of seedlings in a single experiment within 37 days.
Materials and Reagents
- Pipette tips
- General lab materials, including:
Mesh (0.5 mm)
Round Petri dish (9 cm)
Inoculation needle
Microspore glass
Vessel
Funnel
50 ml Falcon tube, etc.
- E. turcicum isolate Passau-1
- Potato dextrose agar (PDA) (BD, DifcoTM, catalog number: 213400 )
- Tween 20 (Sigma-Aldrich, catalog number: V900548 )
- PDA medium (see Recipes)
- Tween 20 solution (see Recipes)
Equipment
- Pipettes
- General greenhouse equipment, including jiffy pots (ø8 cm), tray and sieve tray (L/W: 50 cm/30 cm), etc.
- Home-made iron frame cover with non-permeable plastic (L/W/H: 50/30/35 cm)
- Sprayer (Semadeni, ø28 mm)
- Autoclave
- A home-made box (L/W/H: 54/30/25 cm, open at the bottom, 3 cm notches on each side) to shield the Blacklight Blue fluorescent tubes (Philips TL-D BLB, 15 W, peak at λ 356 nm) or any incubators that can fit the fluorescent tubes can be used alternatively
- Sterile bench with UV light
- Neubauer counting chamber (BRAND, catalog number: 717805 )
- Microscope (ZEISS, model: Axio Imager 2 ) or other light microscopes
- Centrifuge (Eppendorf, model: 5810 R )
Procedure
- Preparation of seedling plants for infection
This seedling assay was conducted under greenhouse conditions (16 h day, 20 °C/8 h night, 18 °C; light intensity, 160 μmol/m2 sec-1 (400 W, MHL); relative humidity, ca. 60%) during the whole experiment. Three to four maize seeds were sown in each jiffy pot, and fifteen pots were placed in one tray (5 per row). After germination, up to 3 seedlings per pot (ca. 45 seedlings per tray) were kept. In general, the second leaves had fully emerged two weeks after sowing. Newly emerged leaves were completely removed by cutting every 2-3 days until the end of each experiment.
Note: Removing the newly emerged leaves can keep seedling plants small and delay leaf senescence, which gives nice disease symptoms and the seedlings can be easily kept under space-limited high-humidity micro-conditions.
- Propagation of E. turcicum isolate
- Inoculation with mycelia and culture on PDA medium plate (35 ml per plate, see Recipes) was carried out at the same day seed sowing (Figure 1A).
- Add 500 ml ddH2O to 19.5 g of PDA powder and autoclave at 121 °C for 20 min. Pour approximate 35 ml of liquid media into round Petri dish plates in a sterile bench. Sterilize the PDA plates under UV light (30 min).
Note: Less media will inhibit fungal growth after some time.
- Inoculation of PDA plates was conducted using an inoculation needle to pick up a small piece of E. turcicum mycelia that is generally kept at room temperature for long-term storage (up to one year). Fix the PDA plate using the permeable surgical tape.
Note: The dehydration and contamination of conidia PDA plates were often observed several months later. Inoculation of PDA plates needs uncontaminated mycelia.
- Place the plates upside down and incubate at room temperature in the dark. When the conidia cover the complete medium plate (after about two weeks), incubate the plates under BLB UV light until harvest (10 h per day, 7 days). This may induce the fungal sporulation to produce more spores.
- Inoculation with mycelia and culture on PDA medium plate (35 ml per plate, see Recipes) was carried out at the same day seed sowing (Figure 1A).
- Harvest and preparation of spore suspension
- Freshly prepare 0.1% Tween 20 (v/v) (see Recipes) in sterile water. Pour 10 ml of 0.1% Tween 20 on the plate and scrape the surface with a glass slide to dislodge the spores from the hyphae.
- Collect and pour the spore suspension by filtering through one layer of a fine mesh (ca. 0.5 mm), which is placed in a funnel to remove most of the mycelia.
- Before counting, mix spore suspension thoroughly by inverting 2-3 times, since the spores settled at the bottom of the collection vessels.
- Determine the spore density by using a 0.1-mm thick Neubauer counting chamber. Adjust the concentration to 4.5 x 104 spores/ml for use. If the spore concentration of the suspension is lower than the concentration needed, concentrate the spores by centrifuging for 5 min (room temperature, 1,811 x g) and removing the extensive supernatant. In general, the spores from one PDA plate after 21 days culture would be enough for infecting 2-4 trays. The conidia PDA plates can be stored up to one year at room temperature, but the spore suspension should be freshly prepared for each infection experiment.
- Freshly prepare 0.1% Tween 20 (v/v) (see Recipes) in sterile water. Pour 10 ml of 0.1% Tween 20 on the plate and scrape the surface with a glass slide to dislodge the spores from the hyphae.
- Infection of maize seedlings by spray
- Inoculate E. turcicum to maize seedlings at 21 days after sowing when the second leaf fully emerges and becomes deep green. This date can be adjusted according to the growth status of seedling plants. Spray four trays using a total of 4 ml of spore suspension.
- After infection, water the tray extensively from the bottom to promote transpiration. A very high humidity micro-environment is achieved by placing non-permeable plastic hoods on top of each tray (Figure 1A).
Note: The high humidity micro-environment can promote E. turcicum infection.
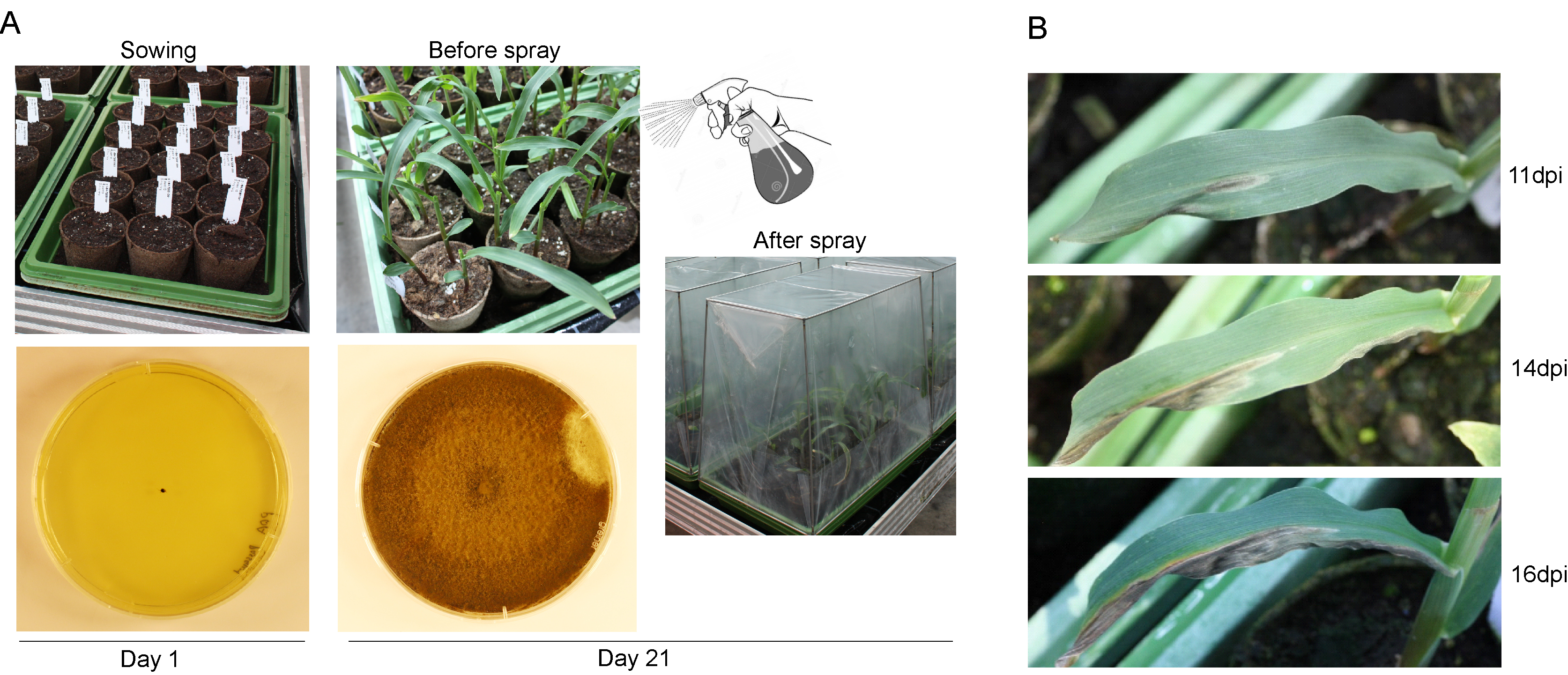
Figure 1. The seedling assay for testing NCLB disease resistance in the greenhouse. A. The pipeline for making inoculation. Parallel sowing and culture start of E. turcicum on a PDA medium plate performed 21 days before inoculation. The first and second leaves were subjected for inoculation by spraying spore suspension (4.5 x 104 spores/ml, 4 ml for 4 trays). The micro-environment with higher humidity was achieved by watering the tray extensively to promote transpiration and by covering the trays with a non-permeable cover until the end of each experiment. B. Symptoms of E. turcicum infected seedling. dpi, days post inoculation.
- Inoculate E. turcicum to maize seedlings at 21 days after sowing when the second leaf fully emerges and becomes deep green. This date can be adjusted according to the growth status of seedling plants. Spray four trays using a total of 4 ml of spore suspension.
- Scoring symptoms
- Scoring individual seedling for disease symptoms was conducted between 11 and 25 days post inoculation in an interval of 1 to 2 days (Figure 1B). The period for scoring can be adjusted according to the levels of susceptibility/resistance.
- Perform scoring for disease symptoms manually by visualization. Three disease parameters AUDPC, PrimDLA at 16 dpi and IP are used for disease quantification. IP and PrimDLA are rated individually, while AUDPC is calculated using the mean of all test plants in each genotype. In case of plants that were uninfected at 25 dpi, the respective IP was rated as 25 days. In general, 10 to 15 plants of each genotype are tested in each experiment. The IP, PrimDLA and AUDPC are calculated as follows:
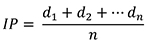
where, d1 = days of symptom appearance post inoculation in plant d1, n = number of total inoculated plants.
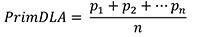
where, p1 = percentage of diseased leaf area of the second leaf in plant p1, n = number of total inoculated plants.
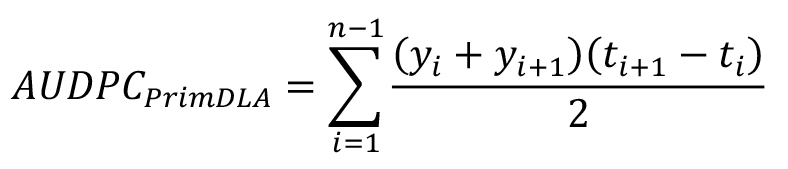
where, yi = PrimDLA at day i, ti+1 - ti = day interval between two ratings, n = number of ratings (Chung et al., 2011).
Note: The use of less plants may lead to large variation and less reliability because of the quantitative nature of NCLB resistance.
- Scoring individual seedling for disease symptoms was conducted between 11 and 25 days post inoculation in an interval of 1 to 2 days (Figure 1B). The period for scoring can be adjusted according to the levels of susceptibility/resistance.
Data analysis
Our previous work demonstrated that a seedling assay can be used to determine the presence of the quantitative NCLB resistance gene Htn1 in maize, as well as the wheat broad-spectrum fungal disease resistance gene Lr34 in transgenic maize lines (Hurni et al., 2015; Sucher et al., 2017). To test if the seedling assay can be used in diverse maize germplasm, we tested this method in a panel of maize lines (Table S1). This panel included six maize breeding lines that were kindly provided by KWS (Einbeck, Germany), and 127 exotic and historic maize lines that were collected before the 1990s from dozens of countries (IPK Genebank, Gatersleben, Germany). Interestingly, a continuous range of NCLB disease resistance/susceptibility was observed (Figures 2A-2C). The susceptible recurrent parental line RP1 was strongly infected, while near-isogenic line containing the introgressed resistance gene Htn1 was highly resistant (Figures 2A-2C). While no visible disease symptoms were detected in ten genotypes, most accessions were infected with visible disease symptoms (AUDPC > 0) and 59% of genotypes were highly susceptible (PrimDLA_16 dpi ≥ 40%). Very importantly, three disease parameters AUDPC, PrimDLA_16 dpi and IP were highly correlated (R2, 81.9 to 94.1) (Figures 2D-2F). Thus, each of the three parameters can be used for quantifying NCLB disease. For example, if PrimDLA is determined at 16 dpi for disease quantification, the data can be obtained after only 37 days.
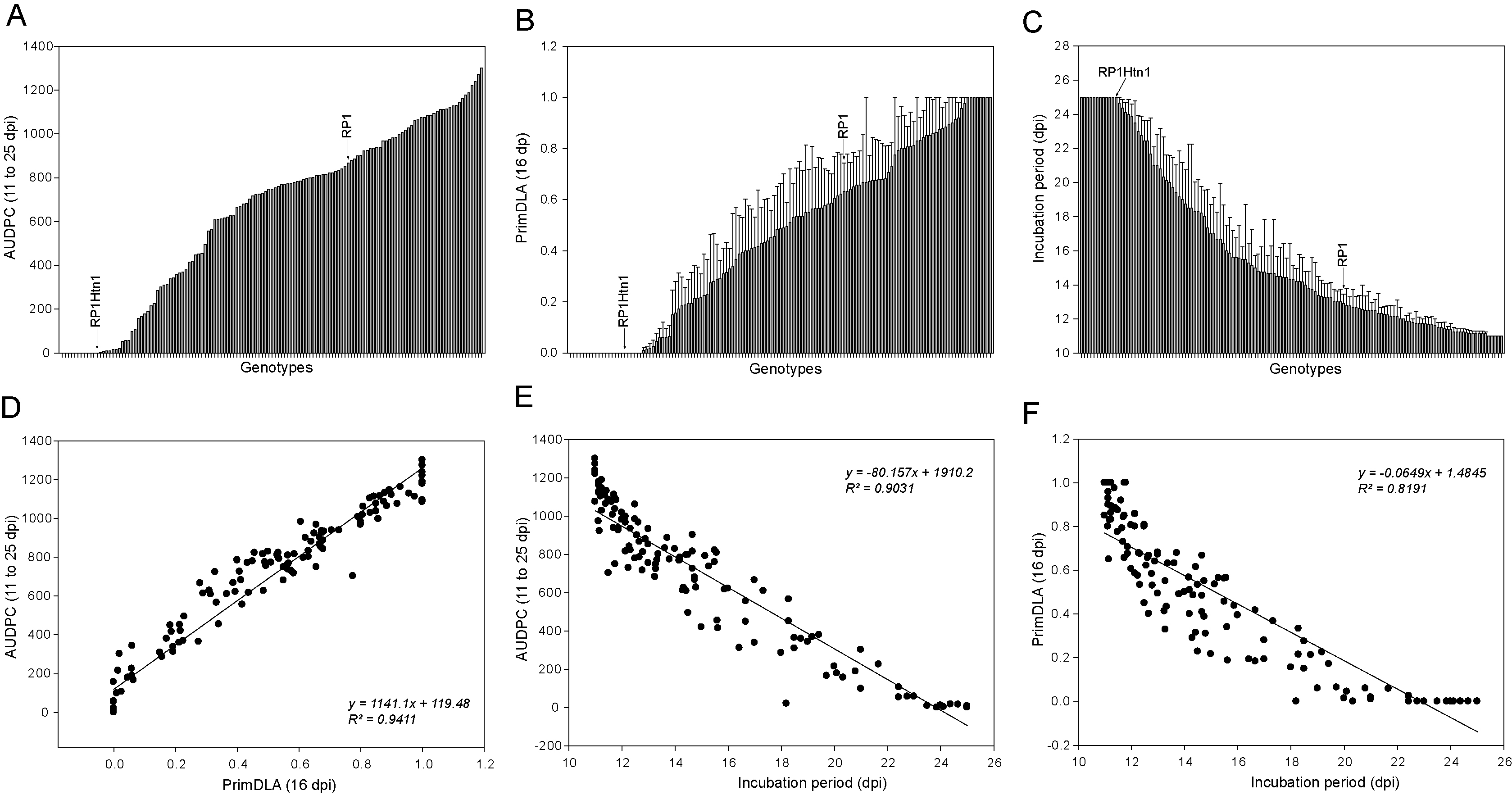
Figure 2. NCLB disease severity and correlation among disease parameters. Six genotypes from KWS Einbeck and 127 exotic maize genotypes from the IPK Genebank were tested in the seedling assay (Table S1). A. The area under the disease progress curve between 11 and 25 dpi; B. The primary diseased leaf (PrimDLA) area at 16 dpi; C. The incubation period that was rated as days from inoculation until appearance of disease symptoms; D. Correlation between AUDPC and PrimDLA at 16 dpi; E. Correlation between AUDPC and IP; F. Correlation between IP and PrimDLA at 16 dpi. Error bars indicate ± standard error (SE). dpi, days after inoculation.
Recipes
- PDA medium
Add 500 ml ddH2O to 19.5 g of PDA powder and autoclave at 121 °C for 20 min
- Tween 20 solution
Add 200 µl Tween 20 to 100 ml of sterile water
Acknowledgments
The authors would like to thank IPK Genebank (Gatersleben, Germany) and KWS (Einbeck, Germany) for kindly providing maize lines, Dr. Severine Hurni for discussion, and Mr. Alessandro Artemisio, Mr. Karl Huwiler for technical support. This work was supported by Swiss National Science Foundation Grant 310030_163260. The protocol was adapted and updated from methods reported in Hurni et al. (2015).
References
- Asea, G., Vivek, B. S., Bigirwa, G., Lipps, P. E. and Pratt, R. C. (2009). Validation of consensus quantitative trait loci associated with resistance to multiple foliar pathogens of maize. Phytopathology 99(5): 540-547.
- Brown, A. F., Juvik, J. A. and Pataky, J. K. (2001). Quantitative trait loci in sweet corn associated with partial resistance to Stewart's wilt, northern corn leaf blight, and common rust. Phytopathology 91(3): 293-300.
- Chung, C. L., Jamann, T., Longfellow, J. and Nelson, R. (2010). Characterization and fine-mapping of a resistance locus for northern leaf blight in maize bin 8.06. Theor Appl Genet 121(2): 205-227.
- Chung, C. L., Poland, J., Kump, K., Benson, J., Longfellow, J., Walsh, E., Balint-Kurti, P. and Nelson, R. (2011). Targeted discovery of quantitative trait loci for resistance to northern leaf blight and other diseases of maize. Theor Appl Genet 123(2): 307-326.
- Dingerdissen, A. L., Geiger, H. H., Lee, M., Schechert, A., Welz, H. G. (1996). Interval mapping of genes for quantitative resistance of maize to Setosphaeria turcica, cause of northern leaf blight, in a tropical environment. Mol Breed 2(2): 143-156.
- Hurni, S., Scheuermann, D., Krattinger, S. G., Kessel, B., Wicker, T., Herren, G., Fitze, M. N., Breen, J., Presterl, T., Ouzunova, M. and Keller, B. (2015). The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc Natl Acad Sci U S A 112(28): 8780-8785.
- Lipps, P. E., Pratt, R. C., Hakiza, J. J. (1997). Interaction of Ht and partial resistance to Exserohilum turcicum in maize. Plant Dis 81(3): 277-282.
- Pataky, J. K., Raid, R. N., Du, T. L. and Schueneman, T. J. (1998). Disease severity and yield of sweet corn hybrids with resistance to northern leaf blight. Plant Dis 82(1): 57-63.
- Sucher, J., Boni, R., Yang, P., Rogowsky, P., Buchner, H., Kastner, C., Kumlehn, J., Krattinger, S. G. and Keller, B. (2017). The durable wheat disease resistance gene Lr34 confers common rust and northern corn leaf blight resistance in maize. Plant Biotechnol J 15(4): 489-496.
- Ullstrup, A. J. (1970). A comparison of monogenic and polygenic resistance to Helminthosporium turcicum in corn. Phytopathology 60(11): 1597-1599.
- Welz, H. G. and Geiger, H. H. (2000). Genes for resistance to northern corn leaf blight in diverse maize populations. Plant Breeding 119: 1-14.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Yang, P., Herren, G., Krattinger, S. G. and Keller, B. (2017). Large-scale Maize Seedling Infection with Exserohilum turcicum in the Greenhouse. Bio-protocol 7(19): e2567. DOI: 10.21769/BioProtoc.2567.
Category
Plant Science > Plant immunity > Host-microbe interactions
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









