- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Capturing Z-stacked Confocal Images of Living Bacteria Entering Hydathode Pores of Cauliflower
Published: Vol 7, Iss 20, Oct 20, 2017 DOI: 10.21769/BioProtoc.2451 Views: 9181
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantification of the Composition Dynamics of a Maize Root-associated Simplified Bacterial Community and Evaluation of Its Biological Control Effect
Ben Niu and Roberto Kolter
Jun 20, 2018 10487 Views

Tracking Root Interactions System (TRIS) Experiment and Quality Control
Hassan Massalha [...] Asaph Aharoni
Apr 20, 2019 7334 Views

A Quick Method for Screening Biocontrol Efficacy of Bacterial Isolates against Bacterial Wilt Pathogen Ralstonia solanacearum in Tomato
Heena Agarwal [...] Niraj Agarwala
Nov 20, 2020 5545 Views
Abstract
The present protocol to visualize living bacteria at the pore level of cauliflower hydathodes is simple and trained users in confocal microscopy can execute it successfully. It can be easily adapted to capture images with other plant-microorganism interactions at the leaf surface and should be useful to obtain important information on pore and stomatal biology. A critical limitation to methods used to observe plant-microorganism interactions in the pore is the application of too much pressure to the sample during observations and z-stack acquisitions. To solve this issue, we recommend the use of a long working-distance water immersion objective lens that allows observations even with thick samples.
Keywords: CauliflowerBackground
Pores of hydathodes and stomata are possible entry points for pathogenic microorganisms to invade plant tissues. In cauliflower, the hydathodes present on leaf margins exhibit large pores, resembling stomata. These pores are routes for the leaf infection by the vascular pathogenic bacterium Xanthomonas campestris pv. campestris (Xcc) (Cerutti et al., 2017). Hydathodes are present on leaves of a wide range of vascular plants. We describe a simple protocol to visualize bacteria at the pore level by confocal microscopy.
Materials and Reagents
- Razor blade
- Microscope glass slides (Thermo Fisher Scientific, Superfrost, catalog number: 10143560WCUT )
- Cover slip 24 x 60 mm (Thermo Fisher Scientific, catalog number: 15747592 )
- Compost (Proveen, catalog number: 14926 )
- Cauliflower (Brassica oleracea var. botrytis, cultivar Clovis, Vilmorin)
Note: Plants were grown on compost in a controlled greenhouse (short day conditions 9 h light; temperature 22 °C; relative humidity 70%). After inoculation, they were placed back in the controlled greenhouse for 24 h inside the miniature greenhouse at 100% relative humidity. After 24 h, lid was removed. All the experiments used the second true leaf from four-week-old plants.
- Xcc strain 8004::GUS-GFP (Xanthomonas campestris pv. campestris (Xcc); Cerutti et al., 2017)
- Magnesium chloride (MgCl2) (Merck, catalog number: 814733 )
- Yeast extract (Sigma-Aldrich, catalog number: Y1625 )
- Casamino acids (BD, BD Biosciences, catalog number: 223050 )
- Potassium phosphate dibasic (K2HPO4) (Merck, catalog number: 105101 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (Merck, catalog number: 105886 )
- Silwet L-77 (CAAHMRO, catalog number: 115950H )
- Calcofluor (Fluorescent Brightener 28) (Sigma-Aldrich, catalog number: F3543 )
- MOKA medium (see Recipes)
Equipment
- Miniature greenhouse purchased from a garden center (Nortene, catalog number: ME673714 )
- Hollow punch (Harris, Uni-Core) (Electron Microscopy Sciences, catalog number: 69039-70 ) or razor blades
- Confocal microscope (Leica Microsystems, model: Leica TCS SP2 or Leica TCS SP8 ) equipped with an argon laser (ray line at 488 nm), a Helium Neon laser at 633 nm and a diode laser at 405 nm
- Water immersion lens 40x with a long working distance (N.A. = 0.8, working distance of 3,300 µm) (Leica Microsystems, catalog number: 50615 5) or water immersion lens 63x with a long working distance (N.A. = 0.9 and a working distance of 2,900 µm) (Leica Microsystems, catalog number: 506148 ) (Leica Mannheim, Germany)
Procedure
- Inoculate plants by dipping leaves (the second true leaf from four-weeks-old cauliflower) for 15 sec inside a bacterial suspension (108 cfu/ml) of 1 mM MgCl2 and 0.02% of Silwet L-77 (see Figure 1A). Bacteria are previously grown in MOKA medium (see Recipe 1). Gently move the leaf in the solution to improve leaf wetting.
- Place the plants for 24 h in closed miniature greenhouses, watered (1-2 cm of water inside the greenhouse) and keep them at 100% relative humidity (see Figure 1B) in a growth chamber. After 24 h, remove lids and place back the plants in the growth chamber (9 h light; 22 °C and relative humidity, 70%).
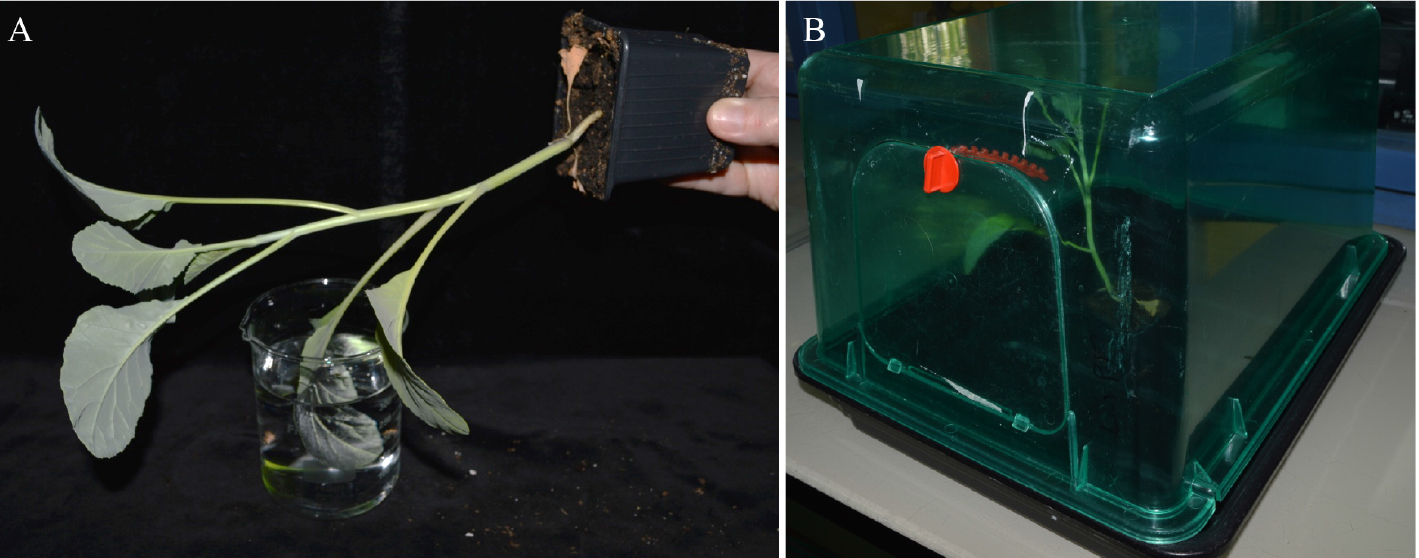
Figure 1. Inoculation procedure of a cauliflower leaf. A. Inoculation by dipping the second true leaf into a bacterial suspension; B. Cauliflower plants are placed after dipping inoculation in miniature greenhouses for 24 h.
- At given days post infection (3 and 6 dpi), take pieces (½ leaf or disk ~0.5 cm in diameter, as shown Figure 2) of leaves (margins including hydathodes) and mount the samples in water on a glass slide (Figure 2). Identify the face to be observed (adaxial, the upper side of the leaf, or abaxial, the underside of the leaf), both sides can be observed. Cover the samples with a cover slip and gently add a few drops of an aqueous solution of calcofluor (0.01%).
Note: Before adding the calcofluor, check the level of autofluorescence at 405 nm. Sometimes the emitted autofluorescence of the epidermal cell wall is sufficient to image the leaf surface.

Figure 2. Samples of cauliflower leaves mounted on glass slide. A. The abaxial face of ½ leaf of cauliflower simply cut with a razor blade; *: Hydathode. The white circle indicates an area cut for observation as shown in B and C. B and C. Leaf disk samples with the hollow punch, with one hydathode (*) at the leaf margin. (B) Adaxial (the upper side of the leaf) and (C) abaxial (the underside of the leaf) views. Scale bars = 2.5 mm.
- To localize regions of interest with bacteria expressing GFP, check the preparation under a microscope (via the eye-pieces) in epifluorescence using a GFP filter cube (excitation in the blue range, and emission filter at 525 ± 25 nm). Use a 10x objective lens to rapidly observe the samples. Then use a 40x or 60x water immersion lenses to acquire images and z-stacks.
- In confocal mode, use the diode laser at 405 nm to collect the fluorescence (between 410-470 nm) emitted by the calcofluor, the 488 nm ray line of the argon laser to collect the GFP fluorescence between 500 and 540 nm and the 633 nm ray line of the HeNe laser to collect the autofluorescence of the chlorophyll (between 640 and 680 nm) (Figure 3).
- Adjust the settings for each channel of fluorescence (power of the laser used for excitation, and the gain of the photomultiplier (PMT) to avoid overexposure of the PMT detectors (Figure 3).
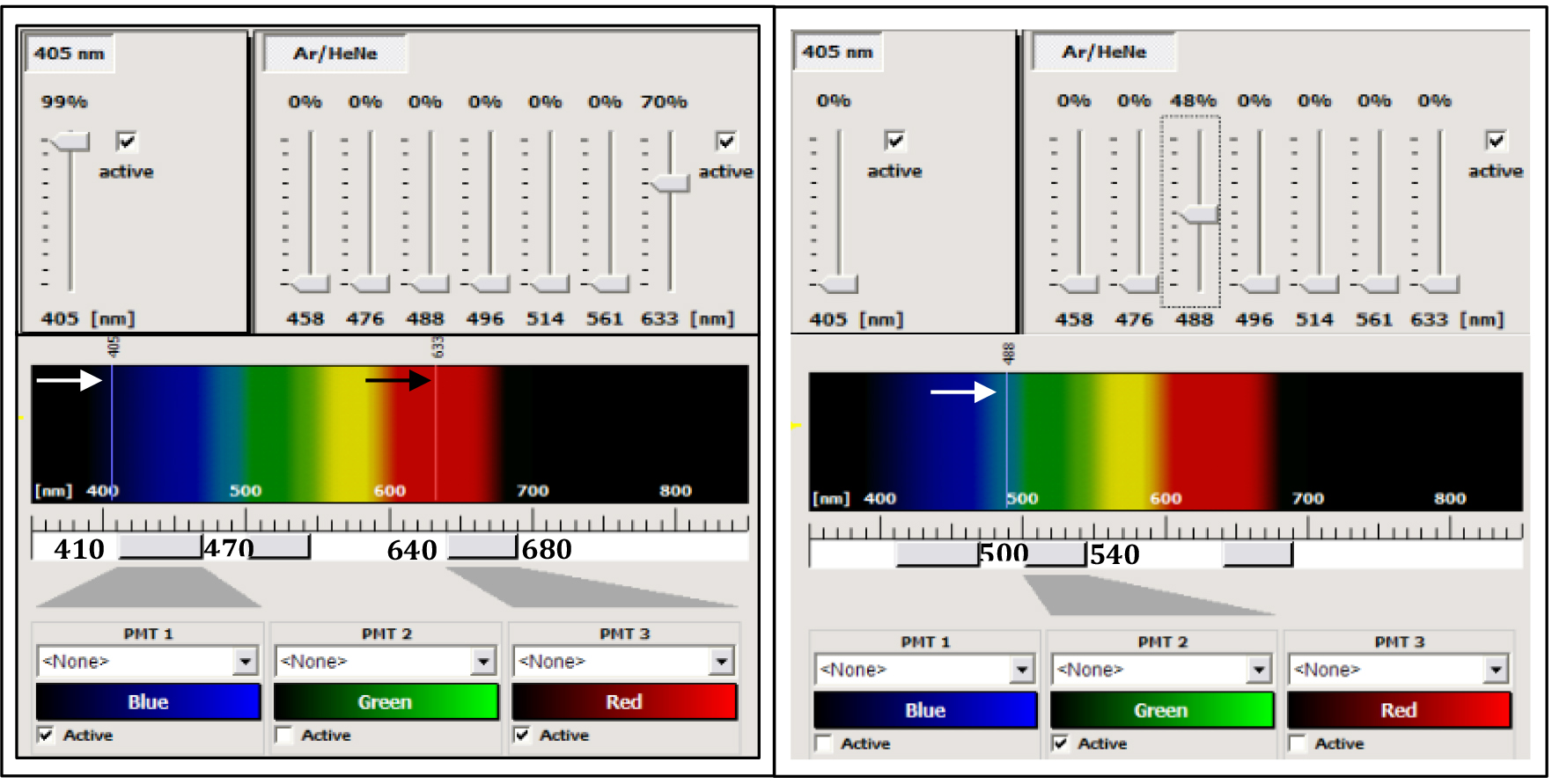
Figure 3. Control panel of the SP2 AOBS confocal microscope. The left part corresponds to the settings for the acquisition of the calcofluor and the chlorophyll fluorescences. The 405 nm ray line (white arrow) of a diode laser is used for excitation of the calcofluor (the power of the laser can be modulated from 1 to 100% of the nominal power of the diode, here the value is 99%). The 633 ray line of a helium-neon laser (black arrow) was used for the excitation of the chlorophyll (power laser at 70% of the nominal power of the laser). The Leica confocal is equipped with a spectral module. The fluorescence emitted by the calcofluor is collected between 410 and 470 nm by the PMT-1 (blue channel), and the fluorescence emitted by the chlorophyll is collected between 640 and 680 nm by the PMT-3 (red channel). The right part corresponds to the settings for the acquisition of the GFP alone, using the 488 nm ray line of an argon laser (white arrow) with a power at 48% of the nominal power of the laser and the fluorescence is collected between 500 and 540 nm by the PMT2 (green channel).
- Use the sequential mode of the confocal microscope to avoid crosstalk between PMT detectors. To do that collect first the calcofluor and the chlorophyll fluorescences and next, the GFP fluorescence as indicated above (Figure 3). Acquire images (512 x 512 pixels) at 400 Hz with a line averaging of 0 to 8 (depending on the speed of bacterial movements) (see Figure 4).
- For z-stack acquisitions, define the top and bottom position of the microscope galvo-stage and the z step close to 0.5 µm. Maximal projection is done (using the Leica Software) from 10 to 25 confocal planes acquired in z-dimension (see Figure 4).
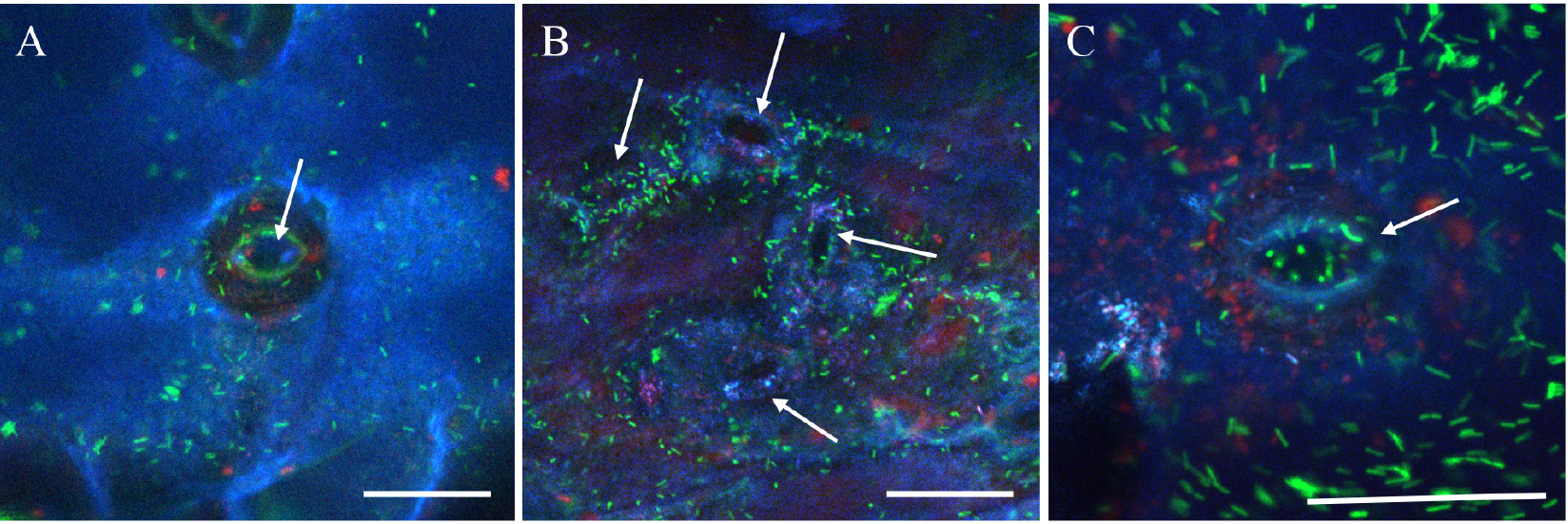
Figure 4. Confocal images of GFP expressing Xcc at the pore level. A. Maximal projection from 15 confocal planes acquired in z-dimension with a z-step of 0.5 µm and a line-averaging value of 2. B. Maximal projection from 20 confocal planes acquired in z-dimension with a z-step of 0.5 µm and a line-averaging value of 4. C. Maximal projection from 25 confocal planes acquired in z-dimension with a z-step of 0.5 µm and no line-averaging. Arrows indicate the pore. The blue color corresponds to the emitted fluorescence of the calcofluor staining the epidermal cell wall surface, the green color corresponds to the GFP expressing bacteria and the red color corresponds to the chlorophyll. Scale bars =30 µm.
Recipes
- MOKA medium
Note: Bacteria were grown overnight at 30 °C in MOKA medium. They were harvested from the culture medium by centrifugation (4,000 x g, 10 min) and suspended in 1 mM MgCl2 and 0.02% of Silwet L-77 at 108 cfu ml-1.
In distilled water for 1 L:
4 g yeast extract
8 g casamino acids
2 g K2HPO4
0.3 g MgSO4·7H2O
Acknowledgments
This work was supported by a PhD grant from the French Ministry of National Education and Research to AC. LIPM is part of the French Laboratory of Excellence project (TULIP ANR-10-LABX-41; ANR-11-IDEX-0002-02). We thank the Région Occitanie for continued financial support of the microscopy platform and Laurent Noël for critically reading the manuscript.
References
- Cerutti, A., Jauneau, A., Auriac, M. C., Lauber, E., Martinez, Y., Chiarenza, S., Leonhardt, N., Berthomé, R. and Noël, L. D. (2017). Immunity at cauliflower hydathodes controls systemic infection by Xanthomonas campestris pv campestris. Plant Physiol 174(2): 700-716.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Cerutti, A. and Jauneau, A. (2017). Capturing Z-stacked Confocal Images of Living Bacteria Entering Hydathode Pores of Cauliflower. Bio-protocol 7(20): e2451. DOI: 10.21769/BioProtoc.2451.
- Cerutti, A., Jauneau, A., Auriac, M. C., Lauber, E., Martinez, Y., Chiarenza, S., Leonhardt, N., Berthomé, R. and Noël, L. D. (2017). Immunity at cauliflower hydathodes controls systemic infection by Xanthomonas campestris pv campestris. Plant Physiol 174(2): 700-716.
Category
Plant Science > Plant cell biology > Cell imaging
Microbiology > Microbe-host interactions > In vivo model > Plant
Cell Biology > Cell imaging > Confocal microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link