- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Tagged Highly Degenerate Primer (THDP)-PCR for Community Analysis of Methane- and Ammonia-oxidizing Bacteria Based on Copper-containing Membrane-bound Monooxygenases (CuMMO)
Published: Vol 7, Iss 12, Jun 20, 2017 DOI: 10.21769/BioProtoc.2354 Views: 8744
Reviewed by: Dennis NürnbergPalash Kanti DuttaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Protocol to Identify Unknown Flanking DNA Using Partially Overlapping Primer-based PCR for Genome Walking
Mengya Jia [...] Haixing Li
Feb 5, 2025 1363 Views
Protocol to Mine Unknown Flanking DNA Using PER-PCR for Genome Walking
Zhou Yu [...] Haixing Li
Feb 20, 2025 1495 Views
Abstract
We describe a two-step PCR strategy using tagged highly degenerate primer (THDP-PCR) targeting copper-containing membrane-bound monooxygenases (CuMMO) genes for community analysis of methane- or ammonia-oxidizing bacteria. This strategy consists of a primary CuMMO gene-specific PCR followed by a secondary PCR with a tag as a single primer. This strategy remarkably increases the divergence of CuMMO gene amplicons while maintaining PCR efficiency without obvious amplification bias. This THDP-PCR strategy can be extended to other functional gene-based community analysis with design of new highly degenerate primer covering target functional gene sequences.
Keywords: Tagged highly degenerate primerBackground
Gene types in CuMMO superfamily are divergent and existent primer sets can only cover some CuMMO types (Tuomivirta et al., 2009). To cover the divergent types of genes in the CuMMO superfamily, highly degenerate primers are inevitable, but previous strategies using highly degenerate primers have limitations when applied to environmental samples, like low PCR efficiency or non-specific amplification (Ledeker and De Long, 2013). We recently used a two-step PCR strategy with tagged highly degenerate primers, designated THDP-PCR, to amplify a wide range of genes in the CuMMO family with satisfactory PCR efficiency and no obvious amplification bias (Wang et al., 2017).
Materials and Reagents
- Pipette tips (20 μl, 200 μl, 1,000 μl) (Thermo Fisher Scientific, Thermo ScientificTM, catalog numbers: 3521-HR , 3551-HR , 3101-05-HR )
- 1.5 ml Eppendorf tubes (Eppendorf, catalog number: T9661-1000EA )
- Strip PCR tubes and caps (Roche Diagnostics, catalog number: 1667009001 )
- PowerSoilTM DNA Isolation Kit (Mo Bio Laboratories, catalog number: 12888-100 )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A7906 )
- Premix Taq (Takara Bio, catalog number: R004A )
- dNTP mixture
- Tris-HCl (pH 8.3)
- Potassium chloride (KCl)
- Magnesium chloride (MgCl2)
- AxyPrepTM PCR Cleanup Kit (Corning, Axygen®, catalog number: AP-PCR-250 )
- Primers

Equipment
- Pipettes (0-10 μl, 10-100 μl, 100-1,000 μl) (Eppendorf, catalog numbers: k03030 , k03031 , k03032 )
- Vortex-genieTM 2 (Mo Bio Laboratories, model: Vortex-Genie® 2 )
- PICO 17 centrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: HeraeusTM PicoTM 17 )
- Automated thermal cycler TP600 (Takara Bio, model: TP600 )
Software
- Mothur (www.mothur.org)
- BLAST+ (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/)
- Megan5 (http://ab.inf.uni-tuebingen.de/software/megan5/)
Procedure
- Sample collection and nucleic acid extraction
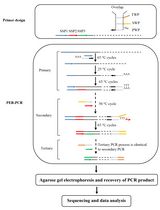
Extract genomic DNA from soil or sediment samples (0.25 g wet weight) using a PowerSoilTM DNA Isolation Kit (Mo Bio Laboratories, USA) according to the manufacturer’s instructions. - THDP-PCR method (Figure 1)

Figure 1. Principle of THDP-PCR- First step of THDP-PCR
- PCR reaction recipe. Final volume: 50 μl
Note: Approximately 2 to 20 ng per sample, H2O for negative control.H2O 18 μl A189-tag (10 μM) 2 μl HD616-tag (10 μM) 2 μl BSA (10 mg/ml) 2 μl Sample DNA 1 μl
Premix Taq (see Table 1) 25 μl
Table 1. Composition of Premix TaqTM solution
Note: Reverse primer HD616-tag was designed based on the CuMMO sequences downloaded from FunGene Pipeline. We aligned the sequences and identified the conserved regions, then introduced degeneracies to cover different types of CuMMO gene sequences, developing the reverse primer HD616. A 20-nt-long tag was added to the 5’ end of both primers, A189 and HD616, to generate the tagged primers A189-tag and HD616-tag, respectively, for THDP-PCR amplification. Tag sequence was from virus genome (Canine astrovirus strain HUN/2012/115, GenBank: KX599351.1) and will not bind to bacterial genomes and thus avoids non-specific PCR amplification. Because tag is used as a single primer in the second step of the THDP-PCR, its length is set to 20 nt, a standard length for primers. - Automated thermal cycler program
1) Denaturation at 94 °C for 5 min
2) 10 cycles at:
94 °C for 1 min
59 °C for 1 min
72 °C for 1 min
3) 72 °C for 10 minNote: Conventional (one step) PCR efficiency is not satisfactory for highly degenerate primers with degeneracy higher than 48 based on our experiment.
- PCR clean-up
Purify the entire volume (50 μl) of the first-step PCR product using an AxyPrepTM PCR Cleanup Kit (Axygen, China) according to the manufacturer’s instructions to remove unbound degenerate primers.
Note: Unbound long tagged 3’-end unmatched degenerate primers will also bind to templates but not trigger the polymerization and thereby prevent the binding of the single tag primer to the first-step PCR products, so PCR clean-up between the two steps is necessary. - Second step of THDP-PCR
- PCR reaction recipe. Final volume: 50 μl
H2O 17-19 μl
Tag-barcode (10 μM) 4 μl
PCR clean-up product 2-4 μl
Note: Including the negative-control sample.
Premix Taq (see Table 1) 25 μl - Automated thermal cycler program
1) Denaturation at 94 °C for 5 min
2) 35-40 cycles at:
94 °C for 1 min
59 °C for 1 min
72 °C for 1 min
3) 72 °C for 10 min
- PCR reaction recipe. Final volume: 50 μl
- Pyrosequencing
Mix together THDP-PCR products from different samples and sequence via Roche_454 pyrosequencing (GS FLX Titanium System) according to the manufacturer’s instructions.
Data analysis
- Trim pyrosequencing reads using the command trim.seqs in mothur with length > 300 and q > 25, and no ambiguous sequences allowed.
- Translate the trimmed sequences into amino acid sequences using FrameBot tools in Functional Gene Pipeline/Repository (http://fungene.cme.msu.edu/) to remove potential non-specific sequences which cannot be translated into CuMMO-related amino acid sequences.
Note: The nucleotide sequences which can pass the translation are retrieved using the command get.seqs in mothur from trimmed sequences, based on this step’s results. The following analysis steps are based on the retrieved nucleotide sequences, not polypeptides. - Identify and remove chimeras using the commands chimera.uchime and remove.seqs in mothur.
- Split the sequences into different samples by the barcode sequence using the command split.seqs in mothur.
- Classify the split sequences with BLAST and the lowest common ancestor method in MEGAN using the expanded CuMMO database.
Note: The original database and parameters used in this step are interpreted by Dumont et al. (2014) in the article and supplementary files. The reference database used in the classification is expanded by Wang et al. (2017) from the original database to suit the broad coverage of the highly degenerate primers, and the detailed information is listed in the article and supplementary files (Wang et al., 2017).
Acknowledgments
This protocol was adapted from our previous studies (Wang et al., 2017). This work was supported by the National Natural Science Foundation of China (NSFC) [grant numbers 31470222, 31170114].
References
- Dumont, M. G., Luke, C., Deng, Y. C. and Frenzel, P. (2014). Classification of pmoA amplicon pyrosequences using BLAST and the lowest common ancestor method in MEGAN. Front Microbiol 5: 34.
- Ledeker, B. M. and De Long, S. K. (2013). The effect of multiple primer-template mismatches on quantitative PCR accuracy and development of a multi-primer set assay for accurate quantification of pcrA gene sequence variants. J Microbiol Methods 94(3): 224-231.
- Tuomivirta, T. T., Yrjala, K. and Fritze, H. (2009). Quantitative PCR of pmoA using a novel reverse primer correlates with potential methane oxidation in Finnish fen. Res Microbiol 160(10): 751-756.
- Wang, J. G., Xia, F., Zeleke, J., Zou, B., Rhee, S. K. and Quan, Z. X. (2017). An improved protocol with a highly degenerate primer targeting copper-containing membrane-bound monooxygenase genes for community analysis of methane- and ammonia-oxidizing bacteria. FEMS Microbiol Ecol 93(3): fiw244.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wang, J., Xia, F., Zeleke, J., Zou, B. and Quan, Z. (2017). Tagged Highly Degenerate Primer (THDP)-PCR for Community Analysis of Methane- and Ammonia-oxidizing Bacteria Based on Copper-containing Membrane-bound Monooxygenases (CuMMO). Bio-protocol 7(12): e2354. DOI: 10.21769/BioProtoc.2354.
Category
Microbiology > Community analysis > THDP-PCR
Molecular Biology > DNA > PCR
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link