- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Extraction and Analysis of Carotenoids from Escherichia coli in Color Complementation Assays
Published: Vol 7, Iss 6, Mar 20, 2017 DOI: 10.21769/BioProtoc.2179 Views: 12233
Reviewed by: Tie LiuBaohua LiBin Tian

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Detection and Quantification of Heme and Chlorophyll Precursors Using a High Performance Liquid Chromatography (HPLC) System Equipped with Two Fluorescence Detectors
Jan Pilný [...] Roman Sobotka
Feb 5, 2015 13026 Views
Heterologous Production of Artemisinin in Physcomitrium patens by Direct in vivo Assembly of Multiple DNA Fragments
Nur Kusaira Khairul Ikram [...] Henrik Toft Simonsen
Jul 20, 2023 2328 Views
Utilizing FRET-based Biosensors to Measure Cellular Phosphate Levels in Mycorrhizal Roots of Brachypodium distachyon
Shiqi Zhang [...] Maria J. Harrison
Jan 20, 2025 2376 Views
Abstract
A common method to investigate the function of genes putatively involved in carotenoid biosynthesis is the so called color complementation assay in Escherichia coli (see, e.g., Cunningham and Gantt, 2007). In this assay, the gene under investigation is expressed in E. coli strains genetically engineered to synthesize potential carotenoid substrates, followed by analysis of the pigment changes in the carotenogenic bacteria via high-performance liquid chromatography (HPLC). Two crucial steps in this method are (i) the quantitative extraction of the carotenoids out of E. coli and (ii) the reproducible and complete separation of the pigments by HPLC.
Here, we present a protocol for the extraction and analysis of carotenoids with a broad range of polarities from carotenogenic E. coli. The solvent mixture used for extraction keeps both the lipophilic carotenes and the more polar xanthophylls in solution and is compatible with the eluent gradient of the subsequent HPLC analysis. The C30-column used is particularly suitable for the separation of various cis-isomers of carotenoids, but also for separation of stereoisomers such as α- and β-carotene or lutein and zeaxanthin.
Materials and Reagents
- 50 ml screwcap polypropylene (PP) tubes, sterile, pyrogen-free (SARSTEDT, catalog number: 62.547.254 )
- 15 ml screwcap PP tubes, sterile (SARSTEDT, catalog number: 62.554.502 )
- Escherichia coli, strain depending on the plasmids used (see Notes)
- Methanol HPLC grade (EMD Millipore, catalog number: 106018 )
- Acetone p.a. grade or higher (EMD Millipore, catalog number: 100014 )
- Dichloromethane p.a. grade or higher (Fisher Scientific, catalog number: 10626642 )
- ddH2O
- Ammonium acetate p.a. grade (EMD Millipore, catalog number: 101116 )
- Acetonitrile HPLC grade (EMD Millipore, catalog number: 113358 )
- Ethyl acetate HPLC grade (Merck Millipore, catalog number: 113353 )
- Lysogeny broth medium (Carl Roth, catalog number: X968.3 )
- Eluents (see Recipes)
- Eluent A
- Eluent B
- Eluent C
Equipment
- For pigment extraction:
- Photometer (Eppendorf, model: Bio Photometer 6131 )
- Vortexer (Scientific Industries, model: G-560E )
- Ultrasonic bath (Elma Schmidbauer, model: Transsonic T 460 , or BANDELIN electronic, model: Sonorex TK52 ) (see Notes)
- Centrifuge (Beckman Coulter, model: Avanti J-25 )
- Centrifuge rotor for 50 ml tubes (Beckman Coulter, model: JA25.50 ) with round bottom adapters for 15 ml tubes (Corning, model: 8441 )
- For HPLC analysis:
- HPLC system with quaternary pump, in-line degasser, thermostatted autosampler, column heater and photodiode-array (PDA) detector (Waters, models: 2795 Separation Module and 2996 photodiode-array detector )
- C30-Column 250 x 4.6 mm (5.0 µm, 250 x 4.6 mm) (Bischoff, model: ProntoSIL 200-5-C30 , catalog number: 2546H300PS050)
- Guard column 20 x 4.0 mm (5.0 µm, 20 x 4.0 mm) (Bischoff, model: ProntoSIL 200-5-C30 , catalog number: 6302H300PS050) connected to column by guard cartridge holder (Bischoff, catalog number: 1502 0508 )
Software
- HPLC-Software (Waters: Empower Pro)
Procedure
- Bacterial culture
- For growth of bacteria, use sterile 50 ml PP tubes (loosely screwed cap) with 15 ml LB-medium for an optimal volume/surface ratio and oxygen supply. Inoculate with freshly transformed E. coli.
- Let the bacterial culture grow at 28 °C and 250 rpm to an OD600 between 0.5 and 1.0. This usually takes between 10 and 16 h, but may vary depending on the carotenogenic plasmid used. If the culture has become too dense, it may be diluted to an OD600 of 0.1 and further incubated until the recommended OD is reached. To avoid photoisomerization of carotenoids accumulating in E. coli, culture growth and pigment extraction should be performed under dim light or red light.
- For growth of bacteria, use sterile 50 ml PP tubes (loosely screwed cap) with 15 ml LB-medium for an optimal volume/surface ratio and oxygen supply. Inoculate with freshly transformed E. coli.
- Cell harvesting and pigment extraction
- Transfer 10 ml of the culture to a 15 ml PP tube, harvest the bacteria by centrifugation (4,000 x g, 4 °C, 4 min) and discard the supernatant by pouring out.
- Spin the bacteria-containing PP tubes again (11,000 x g, 4 °C, 1 min) and remove the remaining supernatant by pipetting. Make sure there is no residual LB medium (Figure 1A gives an example of the resulting bacterial pellet).
- Place the sample on ice to avoid thermal isomerization of carotenoids.
- Add 200 µl prechilled methanol on the pellet and resuspend the cells completely by repeated vortexing (10 sec) and sonication (10 sec) in an ultrasonic bath (see Notes) with intermittent cooling on ice. To avoid warming of the sample, the successive vortexing and sonication cycles should not take longer than 10 sec before putting the tubes back on ice.
- Add 200 µl prechilled acetone and mix by vortexing and sonication as described for step B4.
- Add 200 µl prechilled dichloromethane and mix again by vortexing and sonication as described for step B4.
- Incubate the tube on ice for 2 min. If the (turbid) sample appears homogenous, proceed with step B8; if there is a phase separation, add 50 µl methanol, mix by vortexing and repeat step B7 (see Figure 1 for examples of a homogenous phase [panel B] and unwanted phase separation [panel C] due to incomplete removal of LB medium in step B2).
- Spin the tube (11,000 x g, 4 °C, 4 min) and transfer 400 µl of the colored supernatant containing the solved pigments to an HPLC vial (Figure 1D shows a sample after centrifugation). Make sure not to disturb the pellet.
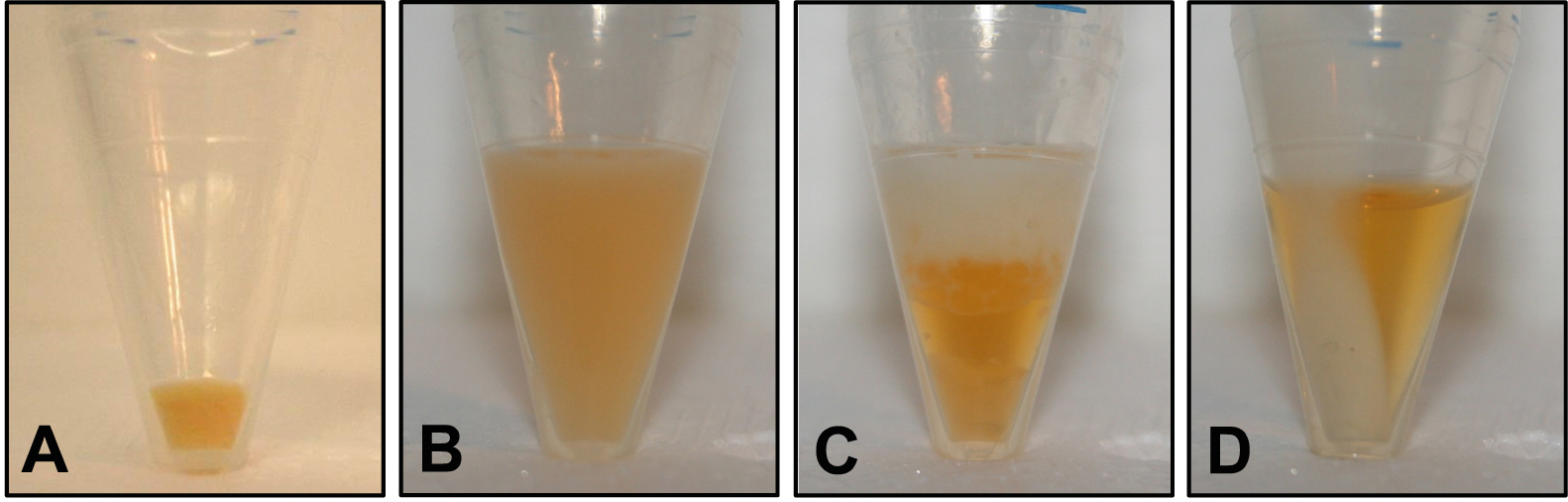
Figure 1. Illustration of different stages of the extraction procedure. A. Bacterial pellet after step B2 (cell harvest by centrifugation); B. Turbid sample resulting in step B7 (extraction of pigments by sonication and vortexing); C. Unwanted phase separation that may occur in step B7 if LB medium was not completely removed in step B4; D. Colored supernatant and colorless pellet after step B8 (clearing extract by centrifugation).
- Transfer 10 ml of the culture to a 15 ml PP tube, harvest the bacteria by centrifugation (4,000 x g, 4 °C, 4 min) and discard the supernatant by pouring out.
- Separation and analysis of carotenoids
- For HPLC analysis, use the following parameters: Column temperature 28 °C; sample temperature 10 °C; injection volume 200 µl; flow 1.3 ml/min, standard detection wavelength 440 nm.
- Before the first measurement, the column should be equilibrated for 10 min at 28 °C with 10% eluent A and 90% eluent B (1.3 ml/min).
- The carotenoids are separated by applying a ternary gradient with linear changes according to Table 1.
- Between successive runs, equilibrate the column for 7 min with 10% eluent A and 90% eluent B (1.3 ml/min). To avoid sample carryover, washing of the injection needle with 100% methanol between HPLC runs is recommended (this is a default procedure of most autosamplers; if you inject manually using a Hamilton syringe, flush the syringe at least three times directly after each sample injection).
- For identification of the most common carotenoids that may be synthesized in color complementation assays using E. coli, compare the retention time and absorbance spectra of the detected pigments with the examples provided here (Figures 2 and 3 in section ‘Representative data’) or use Britton et al. (2004) as reference.
Table 1. Ternary gradient used for HPLC analysis of extracted carotenoids. Eluent composition is given in section ‘Recipes’. The flow is 1.3 ml/min, eluent changes are linear.Time [min] Eluent A [%] Eluent B [%] Eluent C [%] 0 10 90 --- 0.75 --- 100 --- 2.5 --- 60 45 8 --- 50 50 23 --- 10 90 26 --- 10 90 26.5 --- --- 100 30 --- --- 100 32 --- 100 --- 33 10 90 ---
- For HPLC analysis, use the following parameters: Column temperature 28 °C; sample temperature 10 °C; injection volume 200 µl; flow 1.3 ml/min, standard detection wavelength 440 nm.
Representative data
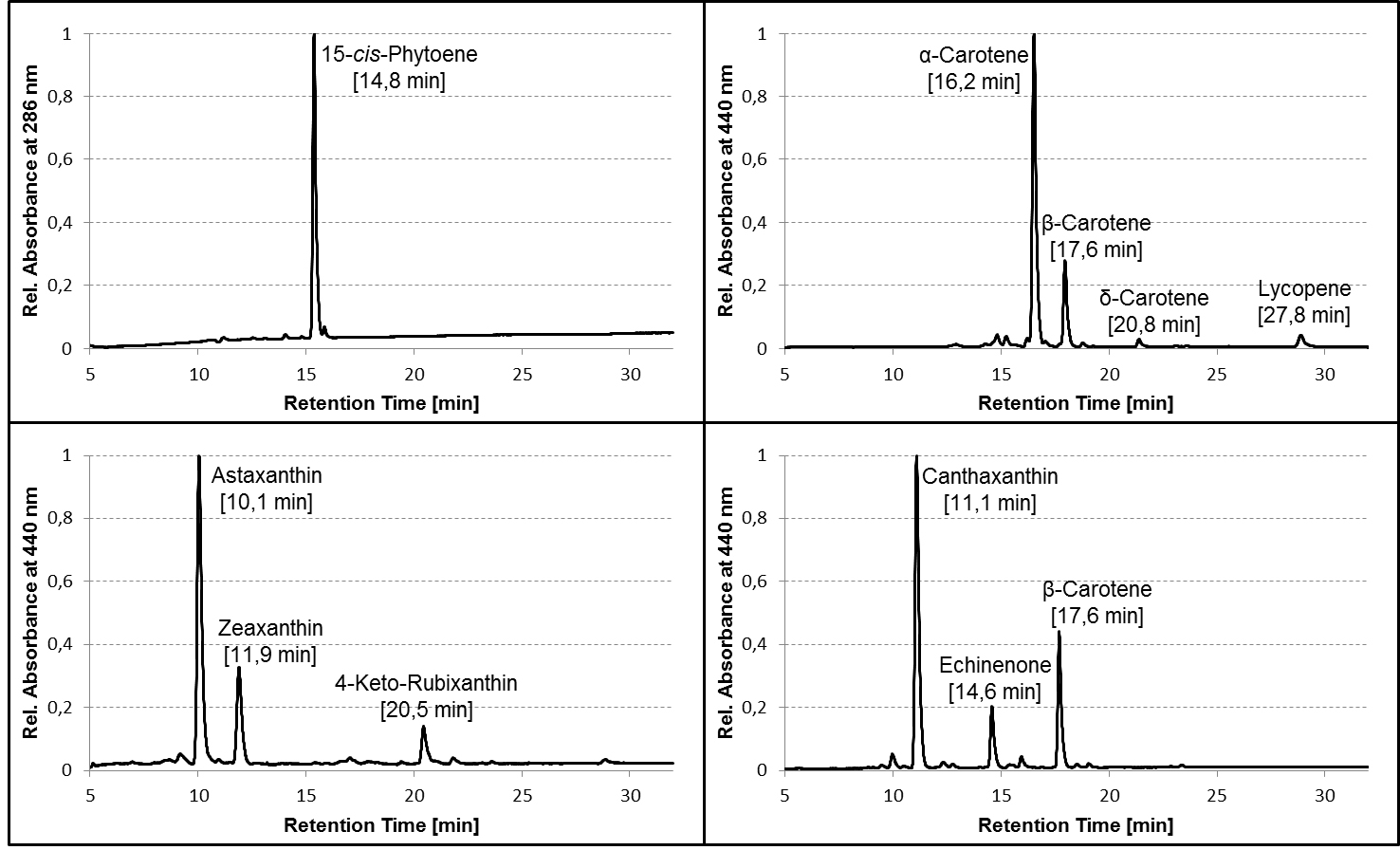
Figure 2. Representative HPLC chromatograms of pigment extracts from carotenogenic strains of E. coli. TOP10 cells were transformed with plasmids containing different combinations of carotenoid biosynthetic genes from bacteria and algae, grown in liquid culture to an OD600 between 0.5 and 1.0, harvested and extracted as described in the protocol, and the pigment extracts analyzed on a C30 column using the HPLC gradient specified in Table 1. Average retention times of the pigments are given in parentheses. Chromatograms are normalized to the peak with the highest absorbance. Note that the relative retention of the pigments (and even their order of elution) may differ considerably between C30 and C18 columns!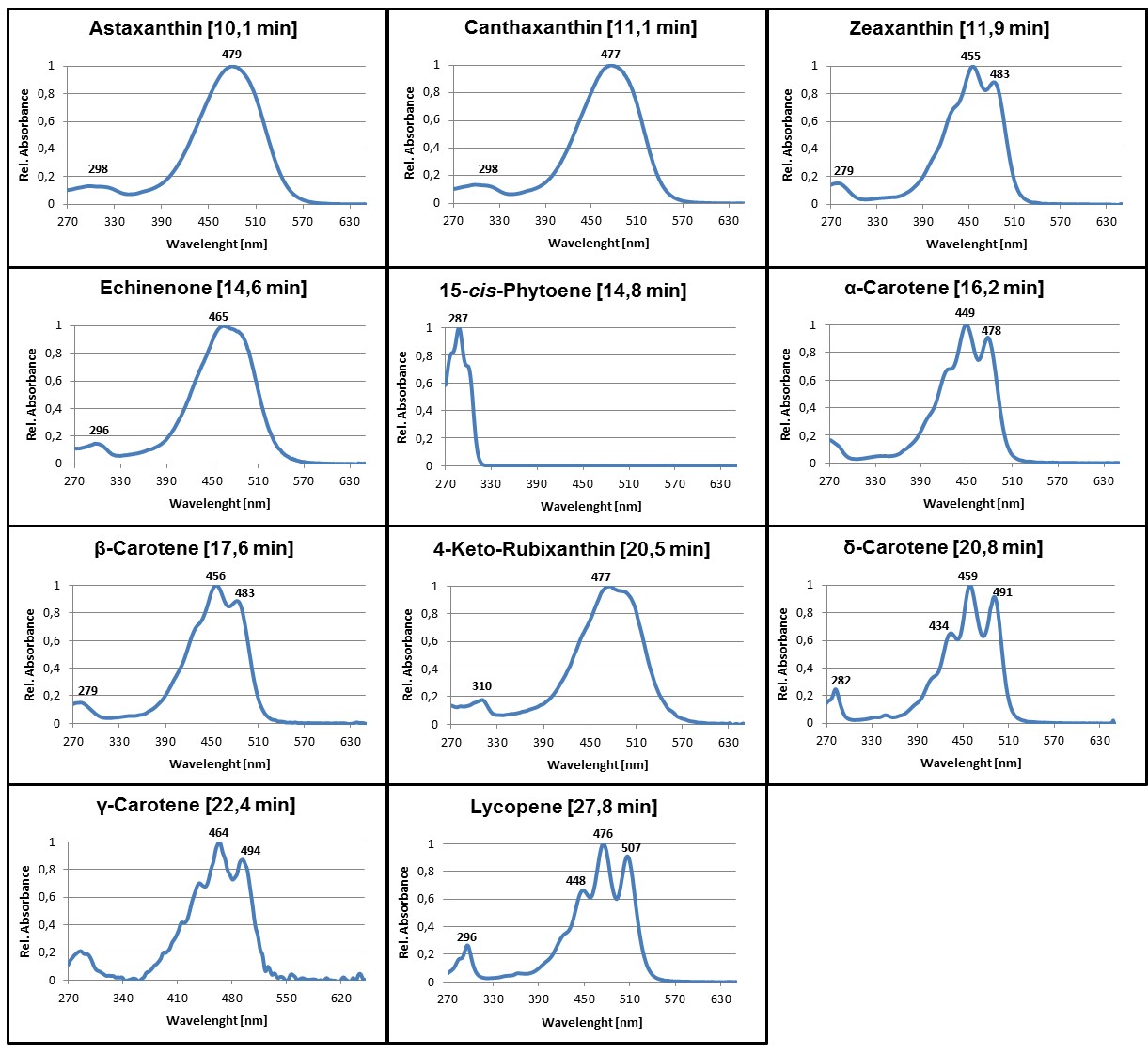
Figure 3. Online PDA spectra of some of the most common carotenoids that may be synthesized in color complementation assays using E. coli. Average retention times of the pigments using a C30 column and the HPLC gradient as specified in Table 1 are given in parentheses. Absorbance spectra are normalized to the maximum absorbance.
Notes
- The choice of a suitable E. coli strain depends on the type of carotenogenic plasmid(s) used and the type of carotenoids that are to be accumulated. Many carotenogenic plasmids are based on pACYC184 (New England Biolabs) as backbone (Misawa et al., 1995; Cunningham and Gantt, 2007). With these plasmids, some authors observed the highest production of carotenoids in TOP10 or TOP10F’ cells (Wurtzel et al., 1997; Cunningham and Gantt, 2007), while others reported contradictory results, recommending SURE cells instead (Chae et al., 2010). We prefer TOP10, because this strain (contrary to most other popular E. coli strains) is not an amber suppressor, thus preventing improper termination of translation for genes with UAG (amber) stop codons. An in depth-discussion on the best combination of strains and plasmids for color complementation assays in E. coli is beyond the scope of this protocol. For an overview on color complementation assays using carotenoid-accumulating strains of E. coli, readers are referred to Cunningham and Gantt (2007).
- Carotenoids are likely to form isomers when exposed to bright light and elevated temperature. To avoid isomerization artifacts, the samples should be protected from light and heat during extraction.
- For sonication, a standard ultrasonic bath (volume of 3 L with a fixed sonication frequency of 35 kHz and between 120 and 240 W power) as used for cleaning procedures is recommended.
- In step B7, phase separation after the addition of dichloromethane only occurs if the extraction solvent is too polar. In most cases, this is caused by incomplete removal of residual LB medium in step B2. Take care to remove all remaining LB medium without disturbing the pellet.
- If the OD of the bacterial cultures at the projected time of harvesting is still low (OD of 0.1-0.3), the amount of solvents used in steps B4-B6 can be scaled down to 100 µl each. In step B8, transfer only 150 µl to the HPLC vial and inject only 100 µl.
- When stored in the autosampler at 10 °C in the dark, the pigments should remain stable and in solution for at least 6 h (we verified this for lycopene, α-carotene and β-carotene).
- Most HPLC methods published for C30 columns apply a binary or ternary gradient using methyl tert-butyl ether (MTBE) instead of ethyl acetate as the strongest eluent. The ternary gradient introduced here is based on eluents commonly used for C18 columns (see e.g., Kraay et al., 1992) and enables switching between C30 and C18 columns without replacement of any eluent. When using a different HPLC gradient, it is critical to verify compatibility of the extraction mixture used in this protocol with the eluent composition applied during sample injection.
- If pigment changes are to be analyzed quantitatively, PDA signals at the detection wavelength must not exceed the linear range of the detector. Otherwise, repeat HPLC analysis with a lower injection volume or a suitable dilution of the respective sample.
Recipes
- Eluents
- Eluent A: 85% (v/v) methanol, 15% ddH2O, 0.075 M ammonium acetate
- Eluent B: 90% (v/v) acetonitrile, 10% ddH2O
- Eluent C: 100% ethyl acetate
Acknowledgments
A short version of this protocol has been published in Blatt et al. (2015). We thank the Carl-Zeiss-Stiftung for financial support.
References
- Blatt, A., Bauch. M. E., Pörschke, Y. and Lohr, M. (2015). A lycopene β-cyclase/lycopene ε-cyclase/light-harvesting complex-fusion protein from the green alga Ostreococcus lucimarinus can be modified to produce α-carotene and β-carotene at different ratios. Plant J 82(4): 582-595.
- Britton, G., Liaaen-Jensen, S., Pfander, H., Mercadante, A. Z. and Egeland, E. S. (2004). Carotenoids Handbook. Birkhäuser.
- Chae, H. S., Kim, K. H., Kim, S. C. and Lee, P. C. (2010). Strain-dependent carotenoid productions in metabolically engineered Escherichia coli. Appl Biochem Biotechnol 162(8): 2333-2344.
- Cunningham, F. X., Jr. and Gantt, E. (2007). A portfolio of plasmids for identification and analysis of carotenoid pathway enzymes: Adonis aestivalis as a case study. Photosynth Res 92(2): 245-259.
- Kraay, G. W., Zapata, M. and Veldhuis, M. J. W. (1992). Separation of chlorophylls c1, c2, and c3 of marine phytoplankton by reversed-phase-C18-high-performance liquid chromatography. J Phycol 28(5): 708-712.
- Misawa, N., Satomi, Y., Kondo, K., Yokoyama, A., Kajiwara, S., Saito, T., Ohtani, T. and Miki, W. (1995). Structure and functional analysis of a marine bacterial carotenoid biosynthesis gene cluster and astaxanthin biosynthetic pathway proposed at the gene level. J Bacteriol 177(22): 6575-6584.
- Wurtzel, E. T., Valdez, G. and Matthews, P. D. (1997). Variation in expression of carotenoid genes in transformed E. coli strains. Biores J 1: 1-11.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Blatt, A. and Lohr, M. (2017). Extraction and Analysis of Carotenoids from Escherichia coli in Color Complementation Assays. Bio-protocol 7(6): e2179. DOI: 10.21769/BioProtoc.2179.
Category
Plant Science > Plant metabolism > Other compound
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











