- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Primer Extension Analysis of HBV DNA with Strand-Specific Primers
Published: Vol 5, Iss 15, Aug 5, 2015 DOI: 10.21769/BioProtoc.1552 Views: 8130
Reviewed by: Smita NairVarpu MarjomakiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Characterization of HBV Isolates from Patient Serum Samples and Cloning
Liang Cao [...] Xinwen Chen
Dec 20, 2015 10499 Views
Quantification of Hepatitis B Virus Covalently Closed Circular DNA in Infected Cell Culture Models by Quantitative PCR
Bingqian Qu and Stephan Urban
Apr 5, 2019 8206 Views
Characterize the Interaction of the DNA Helicase PriA with the Stalled DNA Replication Fork Using Atomic Force Microscopy
Yaqing Wang [...] Yuri L. Lyubchenko
Mar 5, 2021 4777 Views
Abstract
We performed primer extension assay to determine which steps of HBV DNA synthesis (i.e., minus- and plus-strand DNA synthesis and circularization of RC DNA) are affected by phosphoacceptor site mutations in C protein. In these experiments, we used several specific oligonucleotide primers. For quantitation, the level of extended DNA (ED) was normalized to the level of a single internal standard (IS) DNA.
Materials and Reagents
- Huh7 hepatoma cells (Japanese Collection of Research Bioresources Cell Bank, catalog number: JCRB0403 )
- Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies, Gibco®, catalog number: 12800-017 )
- Fetal bovine serum (FBS) (Life Technologies, Gibco®, catalog number: 16000-044 )
- Penicillin/streptomycin (Life Technologies, Gibco®, catalog number: 15140-122 )
- OptiMEM (Life Technologies, Gibco®, catalog number: 31985-062 )
- 500 μl Opti-MEM (Life Technologies, Gibco®, catalog number: 31985-062 )
- PEG (USB, catalog number: 19959 )
- NaCl (Sigma-Aldrich, catalog number: S3014 )
- EDTA (Sigma-Aldrich, catalog number: E5134 )
- Polyethylenimine (Polysciences, catalog number: 23966 )
- Vent Exo (-) polymerase (New England Biolabs, catalog number: M0257S )
- Micrococcal nuclease 1 μl (45 unit/μl) (Worthington Biochemical, I.U.B.: 3.1.31.1, catalog number: LS004798 )
- γ -32P-ATP (PerkinElmer Inc., catalog number: NEG035C )
- T4 polynucleotide kinase (New England Biolabs, catalog number: M0201s )
- Internal standard (IS) DNA (from HBV WT DNA Sac II/Xho I digested fragment) 1ng/1μl
- 2.5 mM dNTP mixture (Takara Bio Company, catalog number: BH7901 )
- RNase A (Fermentas, catalog number: EN05331 )
- Tris-HCl (pH 8.8) (Sigma-Aldrich, catalog number: T6066 )
- (NH4)2SO4 (Sigma-Aldrich, catalog number: T6066)
- KCl (Sigma-Aldrich, catalog number: P9541 )
- MgSO4 (Sigma-Aldrich, catalog number: 230391 )
- Triton X-100 (Sigma-Aldrich, catalog number: T8787 )
- Polyacrylamide (SERVA Electrophoresis GmbH, catalog number: 10687 )
- UREA (Duksan Hi-Metal, catalog number: CAS 57-13-6 )
- APS(Sigma-Aldrich, catalog number: A3678 )
- TEMED (Sigma-Aldrich, catalog number: T9281 )
- Boric Acid(Sigma-Aldrich, catalog number: B0394 )
- EDTA (Sigma-Aldrich, catalog number: E5134)
- 1x DNA-containing reaction buffer (see Recipes)
- 5% polyacrylamide gel (see Recipes)
- 5x TBE (see Recipes)
Equipment
- 10 cm dishes (Corning Incorporated, catalog number: 430167 )
Software
- Fujifilm Image Gauge software (version 4.0)
Procedure
- Huh7 hepatoma cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin under a humidified atmosphere at 37 °C in 5% CO2.
- Cells were passaged every three days. 2 x 106 of Huh7 cells were seeded in 10 cm dish, one day before the transfection.
- Next day, cells were (co)-transfected using polyethylenimine (PEI).
PEI transfection method:
- In a sterile tube, total 10 μg of plasmid DNA (5 μg of phosphoacceptor site mutant and 5 μg of P-deficient mutant) was mixed with 500 μl Opti-MEM.
- Add 30 μl of PEI solution (1 μg/1 μl) to DNA-Opti-MEM solution and then vortex immediately.
- Incubate 15 min at room temperature.
- Then add PEI/DNA-Opti-MEM mixture to cells.
- In a sterile tube, total 10 μg of plasmid DNA (5 μg of phosphoacceptor site mutant and 5 μg of P-deficient mutant) was mixed with 500 μl Opti-MEM.
- Transfection experiments were repeated at least three times.
- Cytoplasmic core particles were prepared as previously described (Kim et al., 2004).
Three days after transfection, cells were used for core particle preparation.
Cytoplasmic core particle preparations:
- Discard medium and wash with 10 ml PBS.
- Add PBS 1 ml, scrape the cells, and transfer to 1.5 ml tube.
- Spin down at 13,000 rpm for 10 sec and discard supernatant.
- Add 1 ml lysis buffer, vortex, and then incubate on ice for 10 min.
- Spin down at 13,500 rpm for 2 min at 4 °C.
- Transfer supernatant to fresh 1.5 ml tube.
- Add micrococcal nuclease 1 μl (45 unit/μl), 1 M MgCl2 10 μl (final 10 mM), 1 M CaCl2 8 μl (final 8 mM).
- Incubate 37 °C, 1 h.
- Add 40% PEG 250 μl (26%), 5 M NaCl 100 μl (1.4 M), 0.5 M EDTA 118 μl (40 mM).
- Put them in ice for 1 h.
- Spin down at 13,500 rpm at 4 °C for 15 min.
- Dissolve pellet in 20 μl nuclease free distilled water.
- Discard medium and wash with 10 ml PBS.
- To analyze HBV DNA synthesis by primer extension analysis, HBV DNA was extracted from isolated core particles (Kim et al., 2004).
Prior to primer extension analysis, HBV DNA synthesis was analyzed by Southern blotting to see the levels of relaxed circular, double-stranded linear, and single-stranded DNAs (Jung et al., 2014) (Figure 1).
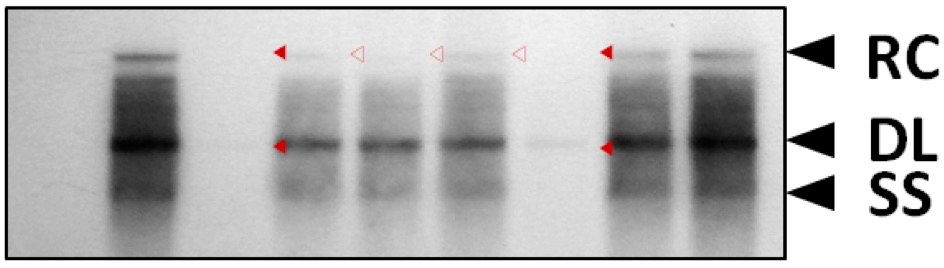
Figure 1. HBV DNA synthesis in core particles formed by STSSSS (WT) and mutant C proteins. To examine HBV DNA synthesis in core particles formed by phosphoacceptor site mutant C proteins, phosphoacceptor site mutant and P-deficient mutant were co-transfected into Huh7 cells. HBV DNA was extracted from isolated core particles (Kim et al., 2004), and Southern blot analysis was performed (Jung et al., 2014). Replicative-intermediate DNAs, relaxed circular (RC), double-stranded linear (DL), and single-stranded (SS) DNAs, are indicated.
- Oligonucleotide DNA primers were 5’-end–labeled with 30 μCi γ-32P-ATP at 37 °C for 3 h using T4 polynucleotide kinase.
Labeling method
The 5’-end–labeled primers HBV1665+ (5’-CTCTTGGACTCTCAGCAATGTCAAC-3’), HBV1744- (5’-CAGCTCCTCCCAGTCCTTAAACA-3’), and HBV1952- (5’- GAGAGTAACTCCACAGTAGCTCC -3’) were used to measure the levels of the elongated minus-strand, plus-strand, and circularized RC DNAs, respectively.Primer 10 pmole/ul
2 μl
γ-32P-ATP
3 μl
D.W
3 μl
10x T4 TNK buffer
1 μl
T4 polynucleotide kinase
1 μl
HBV DNA (total 20 μl) extracted from core particles isolated from co-transfected Huh7 cells in 10-cm dishes was divided into four batches (5 μl): One batch for Southern blotting and three batches for primer extensions to measure minus-strand, plus-strand, and circularized RC DNAs.
For primer extension analyses of each C protein variant, 5ul viral DNA were heated to 95 °C for 5 min, treated with 1 U RNase A at 37 °C for 1 h, ethanol precipitated [ethanol precipitation: To 5 μl extracted DNA, add 195 μl distilled water and 20 μl 3 M sodium acetate, and mix by vortexing briefly. Add 440 μl 100% ethanol (molecular grade), vortex, and keep them overnight at -20 °C. Precipitate DNA by conventional method] and resuspended in distilled water (5 μl). End-labeled primers were extended with Vent Exo (-) polymerase, yielding products that annealed to the respective complementary HBV DNA sequences (Figure 2).
Vortex and spin downViral DNA
5 μl
IS DNA(1 ng/μl)
1 μl
D.W
7 μl
DNA-containing reaction buffer
2 μl
2.5 mM dNTP
2 μl
Vent (-)
1 μl
Labeled primer
2 μl
Total
20 μl
95 °C1 min
95 °C30 sec
60 °C30 sec 20 cycles
72 °C30 sec
72 °C1 min
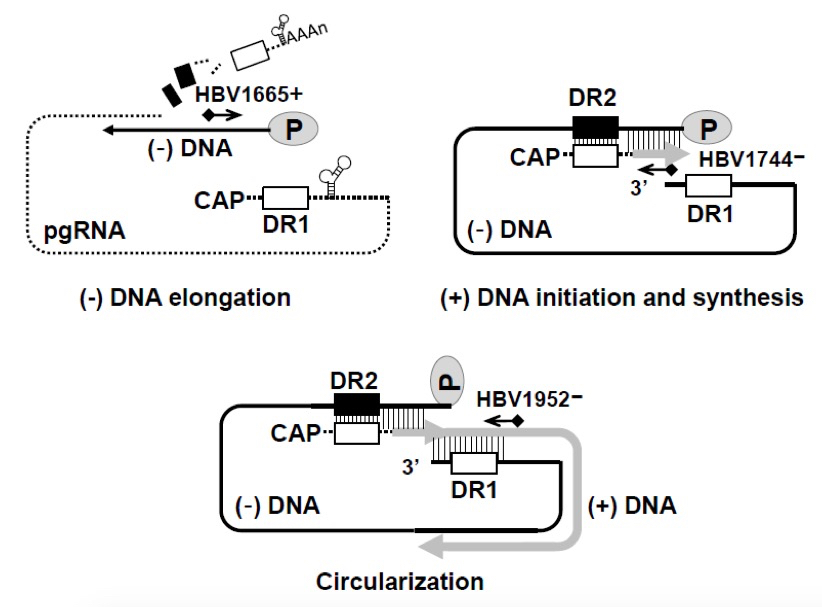
Figure 2. Schematic representation of oligonucleotides used for primer extension analysis. Minus- and plus-strand DNA elongation and circularization were detected using 32P-end-labeled HBV1665+, HBV1744-, and HBV1952-, respectively.
- 5 μl from 20 μl of the extended products were electrophoresed through 5% polyacrylamide gels containing 8 M urea (1,000 voltage 6 h). Gel was dried in gel dryer about 30 min at 60 °C. Dried gels were subjected to autoradiography (Figure 3), and relative levels of radioactivity were measured using the Fujifilm Image Gauge software, version 4.0.
Representative data
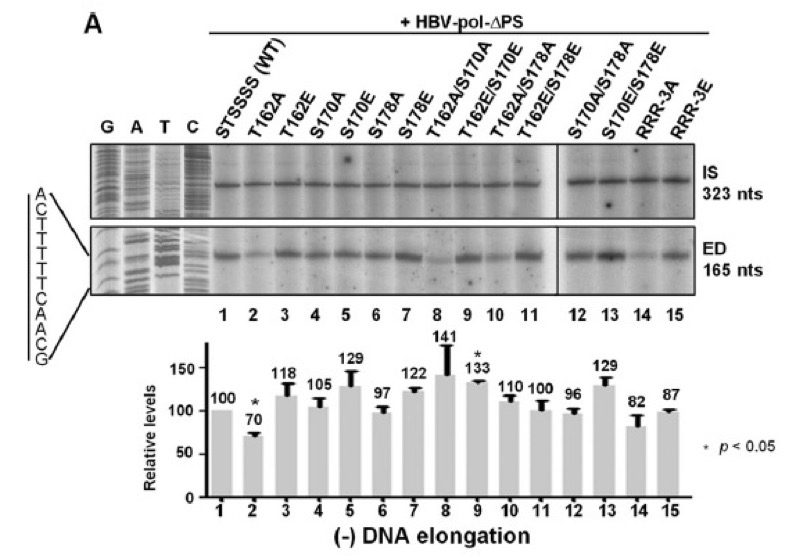
Figure 3. Minus-DNA elongation was detected using 32P-end-labeled HBV1665+, showing that T162A exhibited the reduced minus-strand DNA elongation
Recipes
- Lysis buffer
10 mM Tris (pH 8.0)
1 mM EDTA
0.2% NP40
50 mM NaCl
- 1x DNA-containing reaction buffer
20 mM Tris-HCl (pH 8.8)
10 mM (NH4)2SO4
10 mM KCl
2 mM MgSO4
0.1% Triton X-100
- 5% polyacrylamide gel
30% polyacrylamide
5.2 ml
5x TBE
6 ml
UREA
14.4 g
Add distilled water up to total 30 ml
10% AP
120 μl
TEMED
30 μl
- 5x TBE
Tris-Hcl
54 g
Boric acid
28.5 g
0.5 M EDTA
20 ml
Add distilled water up 1 L
Total
1,000 ml
Acknowledgments
This work was supported by National Research Foundation Grants funded by the Korean Government (NRF-2012-R1A2A2A01015370). We performed primer extension analysis by a previously described method (Lewellyn and Loeb, 2011), with minor modifications.
References
- Kim, H. Y., Park, G. S., Kim, E. G., Kang, S. H., Shin, H. J., Park, S. and Kim, K. H. (2004). Oligomer synthesis by priming deficient polymerase in hepatitis B virus core particle. Virology 322(1): 22-30.
- Jung, J., Hwang, S. G., Chwae, Y. J., Park, S., Shin, H. J. and Kim, K. (2014). Phosphoacceptors threonine 162 and serines 170 and 178 within the carboxyl-terminal RRRS/T motif of the hepatitis B virus core protein make multiple contributions to hepatitis B virus replication. J Virol 88(16): 8754-8767.
- Lewellyn, E. B. and Loeb, D. D. (2011). The arginine clusters of the carboxy-terminal domain of the core protein of hepatitis B virus make pleiotropic contributions to genome replication. J Virol 85(3): 1298-1309.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Jung, J. and Kim, K. (2015). Primer Extension Analysis of HBV DNA with Strand-Specific Primers. Bio-protocol 5(15): e1552. DOI: 10.21769/BioProtoc.1552.
Category
Microbiology > Microbial biochemistry > DNA
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










