- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Genomic DNA Extraction and Genotyping of Dictyochloropsis Green Algae Strains
Published: Vol 5, Iss 15, Aug 5, 2015 DOI: 10.21769/BioProtoc.1545 Views: 13154
Reviewed by: Arsalan DaudiAntoine DanonClaudia Catalanotti

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Safe DNA-extraction Protocol Suitable for Studying Tree-fungus Interactions
Susanna Keriö [...] Jared M. LeBoldus
Jun 5, 2020 7957 Views
Quantification of Botrytis cinerea Growth in Arabidopsis thaliana
Patricia Scholz [...] Athanas Guzha
Aug 20, 2023 2725 Views

Optimized Protocol for DNA Extraction in Three Theobroma Species
Angie F. Riascos-España [...] Pedro A. Velasquez-Vasconez
May 5, 2025 1949 Views
Abstract
Dictyochloropsis is an ecologically important genus of free-living and symbiotic green algae. Representatives of this genus are horizontally transmitted among several fungi of the family Lobariaceae, thus forming photobiont-mediated guilds. This protocol is suitable for extracting DNA from algal cultures and lichen samples and for genotyping seven unlinked Dictyochloropsis reticulata microsatellite markers in a single PCR multiplex.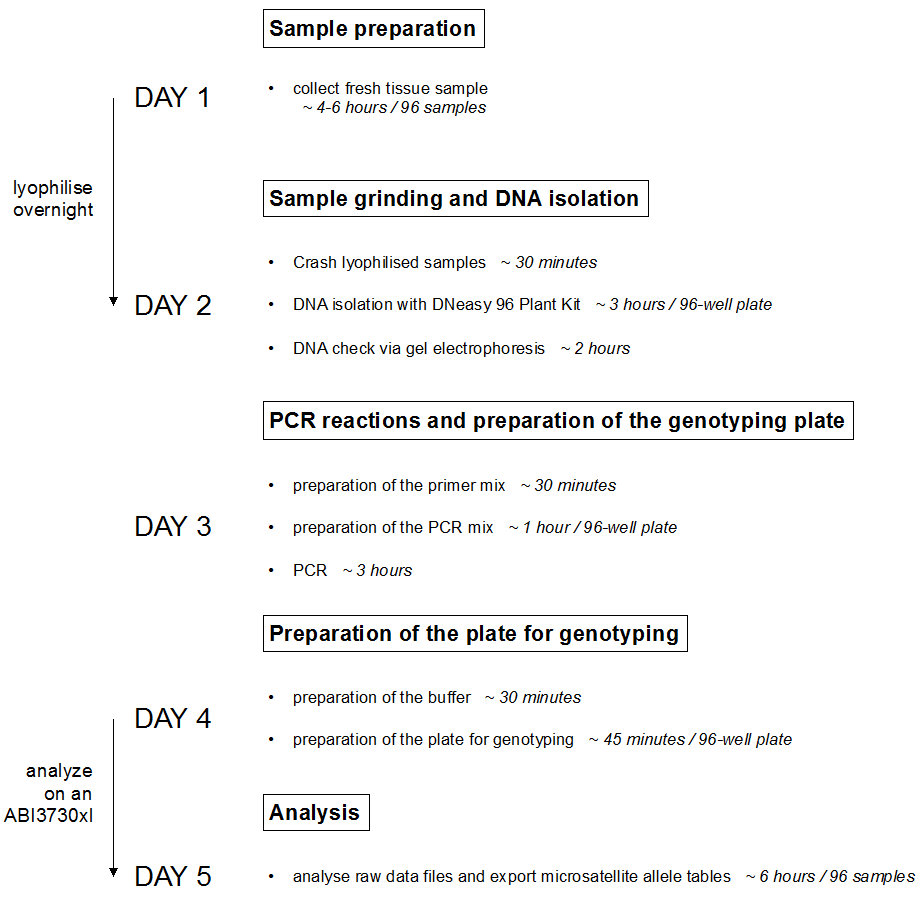
Figure 1. Schematic representation of the analysis pipeline
Materials and Reagents
- Qiagen Type-it Microsatellite PCR kit (QIAGEN, catalog number: 206243 )
- 1x TE buffer (10 mM Tris, bring to pH 8.0 with HCl, 1 mM EDTA)
- GelRed (Biotium, catalog number: 41003 )
- 1 kb DNA ladder (GeneRuler 1 kb DNA Ladder) (Thermo Fisher Scientific, catalog number: SM0313 )
- Nuclease-free water
- Pure molecular biology grade ethanol (96–100%)
- 2 ml tubes
- DNeasy 96 Plant Kit (QIAGEN, catalog number: 69181 )
- Hi-Di™ formamide (Life Technologies, catalog number: 4311320 )
- GeneScan 500 LIZ size standard (Life Technologies, catalog number: 4322682 )
- Ice
- Glucose (BD Biosciences, catalog number: 215510 )
- Proteose pepton (BD Biosciences, catalog number: 212230 )
- Agar (BD Biosciences, catalog number: 214883 )
- 2-[4-(2-hydroxyethyl)-1-piperazinyl]ethanesulfonic acid (HEPES) (Life Technologies, Gibco®, catalog number: 11330032 )
- Laboratory wipes (e.g., KimWipes, Kimberly-Clark, catalog number: TW31KWPBX )
- 4 mm steel balls (e.g., Spex Certiprep, catalog number: 12145950 )
- Pipette tips
- Pipette (0.1-2 μl), pipette tips
- Processing plate: 96-well plate (e.g., MicroAmp Optical 96-Well Reaction Plate) and 96-well plate (Septa)
- 0.22 µm sterile polyethersulfon syringe filter (Merck Millipore, catalog number: P/N SLMP025SS )
- Algal medium (see Recipes)
- Algal culture medium preparation for growing Dictyochloropsis and other trebouxiophycean algal strains (see Recipes)
- Vitamin solution (see Recipes)
Equipment
- Multi-channel pipettes (0.1-10, 10-100, 100-1000 μl), with extended tips
- Microcentrifuge with rotor for 2 ml tubes
- Centrifuge for 96-well plates
- BioRad Gel casting tray, running tray, power pack etc. (Bio-Rad Laboratories, catalog number: 164-0305 )
- Incubator (65 °C)
- Freezer or cold room at -20 °C
- PCR Thermal cycler
- ABI3130/ 3130xl/3730/3730xl DNA Analyzer (Applied Biosystems)
- Ball Mill MM 400 (Retsch®, catalog number: 20.745.0001 )
- Lyophilizer (e.g., FreeZone 4.5 Liter Benchtop Freeze Dry System, Labconco, catalog number: 7750021 )
Software
- GeneMapper® (Software v4.1, Applied Biosystems, catalog number: 4366925)
Procedure
- Sample preparation (Day 1)
- Take up to 10 mg fresh tissue sample (approximate wet weight) or 5 mm diameter
in size of algal culture (Beck et al., 1998) / lichen thallus in 2 ml tube (see Figure 2).
- Add 2 steel balls in each tube.
- Cover the open tubes with soft tissue paper.
- Cool samples in -20 °C freezer for at least 6 h.
- Fit the tubes in the lyophilizer and lyophilize overnight.
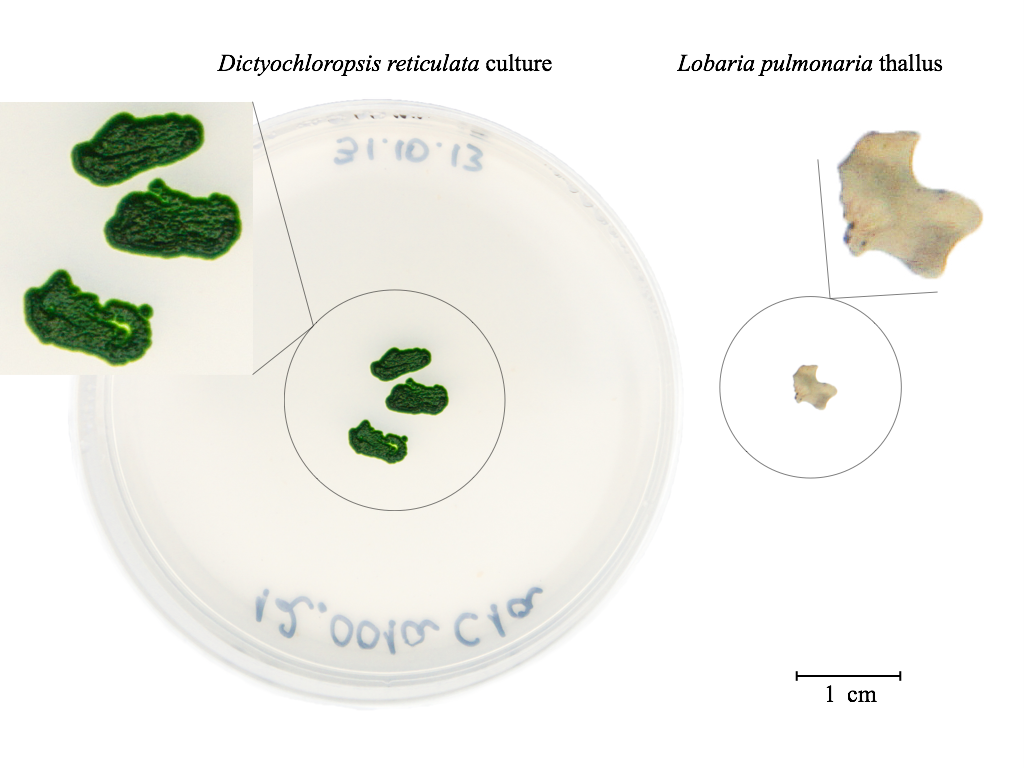
Figure 2. Example of samples used for DNA extraction
- Take up to 10 mg fresh tissue sample (approximate wet weight) or 5 mm diameter
in size of algal culture (Beck et al., 1998) / lichen thallus in 2 ml tube (see Figure 2).
- Sample grinding and DNA isolation (Day 2)
- Close the tubes.
- Crash the lyophilized samples using the ball mill MM 400 at 30 Hz for 30 sec without buffer at ambient temperature.
- Spin down for a few seconds the crashed sample before proceeding with DNA isolation.
- For DNA isolation follow DNeasy 96 Plant Kit, Qiagen protocol (Plant Handbook 10/2012, http://www.qiagen.com, pages 31-34).
Notes:
- For buffers AW1 and AW2, before using for the first time, add the appropriate amount of ethanol as indicated on the bottle to obtain a working solution.
- Preheat Buffer AP1 to 65 °C.
- For buffers AW1 and AW2, before using for the first time, add the appropriate amount of ethanol as indicated on the bottle to obtain a working solution.
- Elute the purified DNA from the DNeasy spin column using Buffer AE twice (2 x 50 μl).
Note: Smaller or larger elution volumes can be used for more or less concentrated products. To ensure complete elution, 40 μl should be the minimum elution volume.
- Close the tubes.
- DNA check
- Prepare 1% agarose gel with 1x TAE buffer. Add 0.5 μl GelRed for 100 ml gel.
- Load 2 μl DNA.
- Add 1 kb or 100 bp ladder for reference.
- Run the gel electrophoresis.
- Check DNA for quality and quantity in gel doc system (Figure 3).
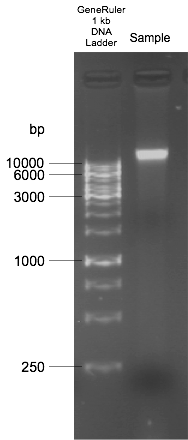
Figure 3. Example of good quality genomic DNA of Dictyochloropsis reticulata run on a 1% w/v agarose gel
- Prepare 1% agarose gel with 1x TAE buffer. Add 0.5 μl GelRed for 100 ml gel.
- PCR reactions and preparation of the genotyping plate (Day 3)
7x Primer mix
- First prepare 100 μM primer stocks.
- Mix all primers according to Tables 1 and 2 to get 1ml primer-mix (sufficient for 10x 96-sample PCR plate).
Table 1. Primer sequences and labeling of the microsatellite Multiplex 1 specific to Dictyochloropsis reticulata (Dal Grande et al., 2009; Dal Grande et al., 2014)
Algal Loci
Locus GenBank
Primer Sequence (5'-3')
Label
LPh1
FJ754261
F: GTCTCAGGTGACCACTTGATTG
VIC
R: GCAATGGATATGATGCTTGTTC
LPh2
FJ754262
F: GACAGCTGTTCCAGTGCATC
R: GCAGAGGAAGTGCATGACG
FAM
LPh3
FJ754263
F: TGCAGTAGGTGTCATATGTGT
NED
R: GAAGGCGCATCTTGATATAC
LPh4
FJ754264
F: GTGGTGGTACAACATGCTCA
NED
R: ACGACCACGTGGGATATCTA
LPh5
FJ754265
F: TGGTGTTAGTAAGAATCGGCATC
PET
R: GTGTATGTCGGCCCCAATAA
LPh6
FJ754266
F: GAATCCTGCCTGCCTACAAG
FAM
R: AGCAACCCATTTCAACCAAC
LPh7
FJ754267
F: TGTGACAGGTGAAACACCAA
VIC
R: TATGGTCCCTCATGGCAAAT
Table 2. 7x primer mix preparation
Concentration of normalized primer stocks: 100 μM (100 pmol/μl)
Each primer 20 μl (30 μl for LPh7)
TE buffer 700 μl
Total volume 1 ml
- First prepare 100 μM primer stocks.
- PCR reaction mix
- Thaw template DNA, RNase-free water, the primer mix and the 2x Type-it Microsatellite PCR Master Mix, if stored at -20 °C.
- Mix the solutions completely before use.
- Prepare a reaction mix according to Table 3.
Note: The reaction mix contains all the components required for multiplex PCR except the template DNA. Prepare a volume of reaction mix 10% greater than that required volume for the total number of reactions to be performed.
- Mix the reaction mix thoroughly and dispense appropriate volumes into PCR tubes or plates.
Note: Mix gently by pipetting the reaction mix up and down a few times. It is not necessary to keep samples on ice during reaction setup.
Table 3. PCR components of the microsatellite multiplex (total volume = 10 μl)
PCR component
Multiplex 1 (10 μl)
Multiplex PCR Master Mixa
5 μl (1x)
Primer mix
1 μl (0.2 μM of each primer, 0.3 μM for LPh7)
DNA template
1 μl (1–10 ng)
RNase-free water
3 μl
- Thaw template DNA, RNase-free water, the primer mix and the 2x Type-it Microsatellite PCR Master Mix, if stored at -20 °C.
- PCR program
- Step 1: 95 °C for 5 min (initial activation)
- Step 2: 95 °C for 30 sec (denaturation)
- Step 3: 62 °C for 90 sec (annealing)
- Step 4: 72 °C for 60 sec (extension)
- Step 5: go to step F2, repeat 24 times
- Step 6: 60 °C for 30 min (final extension)
- Store PCR products in -20 °C, until further processing.
- Step 1: 95 °C for 5 min (initial activation)
- Preparation of the plate for genotyping (Day 4)
- Dilute the PCR products 1: 10 with RNAse-free water.
- Combine 1 μl of diluted PCR product with a buffer containing 9 μl of a denaturing agent (Hi-Di™ Formamide) and 0.5 μl LIZ500 size standard.
- Note: For one 96 well plate prepare buffer for 105 samples.
- Centrifuge the plates briefly (5 sec) at 1,500 RCF.
- Prepare table of samples.
Note: Fill any blank sample-well with nuclease-free water.
- Analyze on an ABI3730xl.
- Dilute the PCR products 1: 10 with RNAse-free water.
- Analysis (Day 5)
Import the raw data files generated by the sequencer to the computer with GeneMapper software.
Genotyping in GeneMapper
- Select ‘Create new project’ from the 'File' menu.
- Import your samples.
- Create marker panel and bin set.
Analysis parameters
- Choose ‘Microsatellite default’ as table setting and ‘Microsatellite analysis method’ as the analysis method.
- Set the panel for the 7x-primer mix.
- Set size standard as LIZ500 and exclude the 35- and 250-bp peaks.
- Analyze (see Figure 4).
Note: If some samples fail to match size standards, go to edit size standards and override size. If this does not solve the issue, there might be some problem with the sample or the analysis run.
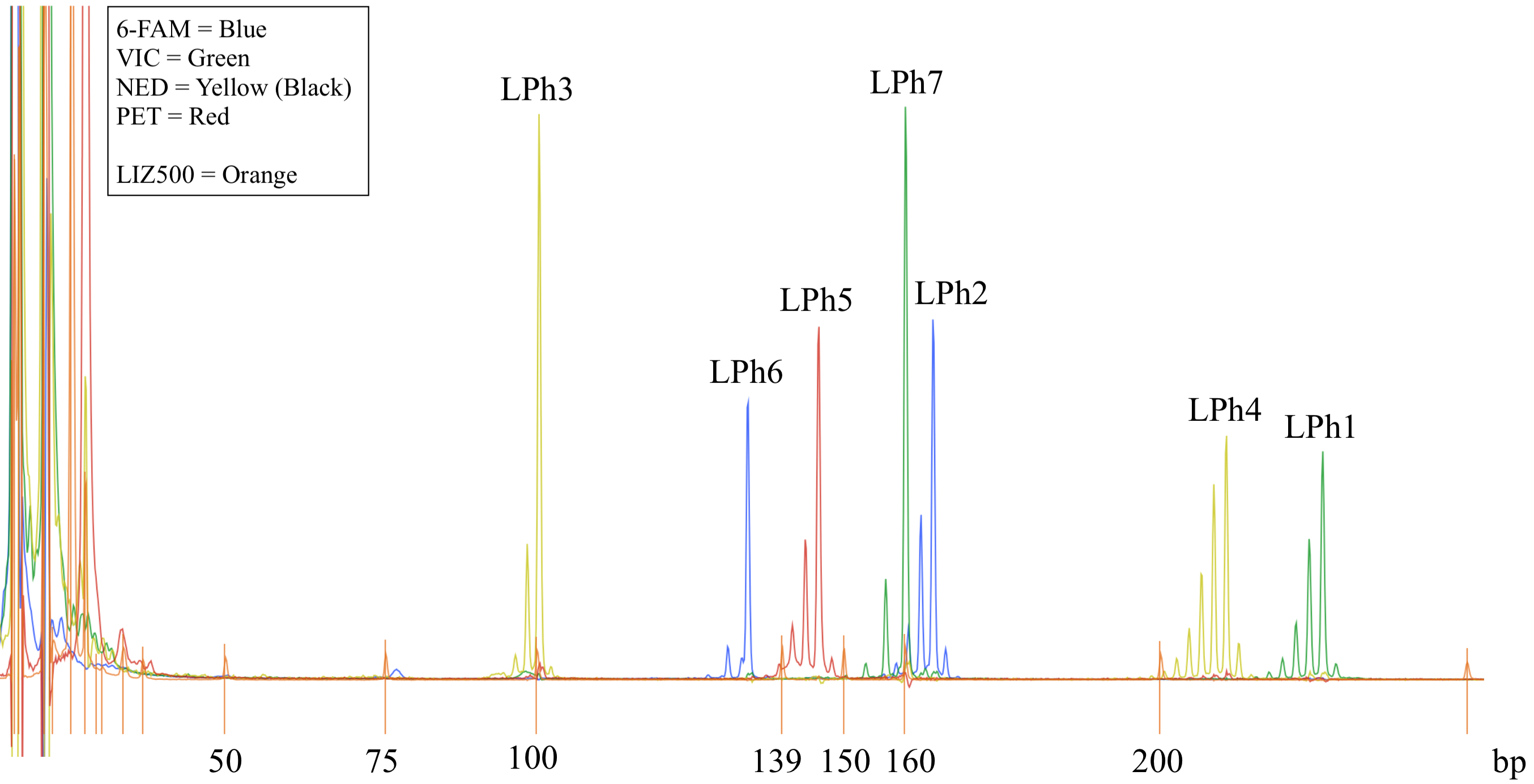
Figure 4. Example of the 7x-multiplex microsatellite electropherogram for one sample of Dictyochloropsis reticulata
- Select ‘Create new project’ from the 'File' menu.
Representative data
- A simple, representative example of data that indicates what type of results to expect (Table 4).
Table 4. Example of a 7x-multiplex microsatellite allele table
Note: Numbers indicate the relative size of each microsatellite allele in base pairs.Sample
Population
LPh1
LPh2
LPh3
LPh4
LPh5
LPh6
LPh7
1
Pop1
262
157
108
191
143
121
170
2
Pop2
260
151
138
176
156
121
190
3
PopN
148
159
122
181
140
116
152
Recipes
- Algal medium
Ingredients for the preparation of algal culture medium
- Macronutrients (g/400 ml)
NaCl 1 (42.7 mM)
CaCl2.2H2O 1 (17.0 mM)
KNO3 20 (495 mM)
MgSO4.7H2O 3 (30.4 mM)
(NH4)2.HPO4 10 (189 mM)
- Micronutrients (g/500 ml)
KOH 15 (535 mM)
EDTA: 25 (171 mM)
FeSO4.7H2O 2.49 (17.9 mM)
H3BO3 5.52 (179 mM)
ZnSO4.7H2O 4.41 (30.7 mM)
MnCl2.4H2O 0.72 (7.28 mM)
NaMoO4 0.36 (3.50 mM)
CuSO4.5H2O 0.79 (6.33 mM)
Co(NO3)2.6H2O 0.25 (1.72 mM)
- Macronutrients (g/400 ml)
- Algal culture medium preparation for growing Dictyochloropsis and other trebouxiophycean algal strains
- For one liter of medium, take 10 ml of macronutrients and 1 ml of micronutrients, add 0.715 g HEPES buffer (final concentration 3 mM) and fill up to 1,000 ml with distilled water.
- Adjust pH to 5.5 with HCl.
- 20 g agar may be added.
- Autoclave and add 1 ml sterile vitamin solution after cooling down to at least 60 °C. Sterilize the vitamin solution by filtration using a 0.22 µm sterile polyethersulfon syringe filter.
Note: To receive higher amounts of algal cells 1.5% glucose and 1% proteose pepton can be added.
- For one liter of medium, take 10 ml of macronutrients and 1 ml of micronutrients, add 0.715 g HEPES buffer (final concentration 3 mM) and fill up to 1,000 ml with distilled water.
- Vitamin solution
Thiamine 0.1 g/100 ml (3.77 mM)
Biotin 2.5 mg/100 ml (0.10 mM)
Vitamin B12 1.5 mg/100 ml (9.50 µM)
Acknowledgments
This study was supported by ‘LOEWE, Landes-Offensive zur Entwicklung Wissenschaftlich-oekonomischer Exzellenz’ of Hesse’s Ministry of Higher Education, Research, and the Arts, by the Swiss National Science Foundation (projects 31003A-105830 and 31003A-127346 to C.S.), and by the German National Science Foundation (project BE3825/4-1 to A.B.).
References
- Beck, A., Friedl, T. and Rambold, G. (1998). Selectivity of photobiont choice in a defined lichen community: inferences from cultural and molecular studies. New Phytologist 139(4): 709-720.
- Dal Grande, F., Beck, A., Cornejo, C., Singh, G., Cheenacharoen, S., Nelsen, M. P. and Scheidegger, C. (2014). Molecular phylogeny and symbiotic selectivity of the green algal genus Dictyochloropsis s.l. (Trebouxiophyceae): a polyphyletic and widespread group forming photobiont-mediated guilds in the lichen family Lobariaceae. New Phytol 202(2): 455-470.
- Dal Grande, F., Widmer, I., Beck, A. and Scheidegger, C. (2010). Microsatellite markers for Dictyochloropsis reticulata (Trebouxiophyceae), the symbiotic alga of the lichen Lobaria pulmonaria (L.). Conservation Genetics 11(3): 1147-1149.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Grande, F. D., Cornejo, C., Scheidegger, C. and Beck, A. (2015). Genomic DNA Extraction and Genotyping of Dictyochloropsis Green Algae Strains. Bio-protocol 5(15): e1545. DOI: 10.21769/BioProtoc.1545.
Category
Plant Science > Phycology > DNA > Extraction
Plant Science > Plant molecular biology > DNA > DNA extraction
Molecular Biology > DNA > Genotyping
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










