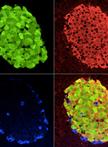
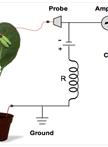
Alkaline Phosphatase Staining
Two main features characterize pluripotent cells; self-renewal (unlimited cell division) and the ability to give rise to all cells of the adult organism. Given the recent impact of induced pluripotent stem cells (iPSCs) and ongoing use of pluripotent embryonic stem cells ESCs (ESCs) in basic discovery, drug development, and potential use for stem cell therapy and regenerative medicine, methods to definitively distinguish pluripotent cells from their differentiated derivatives are required. This will allow us to better understand the factors that promote their survival, self-renewal, and lineage-specific differentiation. Undifferentiated embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) may be identified through the use of biomarker and functional assays. Biomarker assays include those for transcript and protein expression of important pluripotency transcription factors (OCT4, SOX2, and NANOG), cell surface markers (SSEA-1, -3, and -4; TRA-1-60, TRA-1-81), and Alkaline Phosphatase (AP) activity (Brambrink et.al., 2008; Ginis et al., 2004). Functional assays include: (1) the ability to generate teratomas consisting of cells from all three germ layers (endoderm, ectoderm, and mesoderm) when transplanted into immunodeficient mice or upon in vitro differentiation; (2) the ability to generate a chimera; and (3) germline transmission (Marti et al., 2013; Buehr et al., 2008). The latter two tests are ethically feasible only for mouse and other non-human pluripotent cells. In this protocol (Campbell and Rudnicki, 2013) we describe a rapid method to screen for pluripotent cells by AP activity. AP, also known as Basic Phosphatase catalyzes the dephosphorylation of many molecules including nucleotides and proteins. AP activity is high in pluripotent cells but is greatly decreased in more differentiated cell types. The technique described herein may be used to enumerate pluripotent cells during differentiation in the presence or absence of specific genetic manipulations or small chemical modulators. It may also be used to monitor induced pluripotency using defined factors from more differentiated cell types.