- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2016
Volume: 6, Issue: 21
Biochemistry
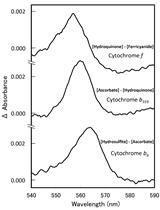
Quantitation of Cytochromes b559, b6, and f, and the Core Component of Photosystem I P700 in Cyanobacterial Cells
Cancer Biology
Sulforhodamine B (SRB) Assay in Cell Culture to Investigate Cell Proliferation
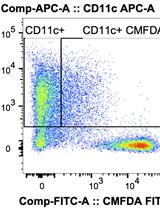
Phagocytosis Assay to Measure Uptake of Necroptotic Cancer Cells by BMDCs
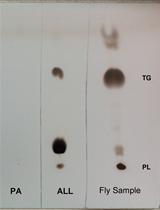
Fat Turnover Assay in Drosophila
Cell Biology
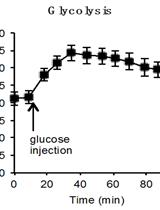
Determination of the Glycolysis and Lipogenesis in Culture of Hepatocytes
Immunology
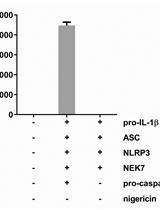
Reconstruction of the Mouse Inflammasome System in HEK293T Cells
Microbiology
Sequencing of Ebola Virus Genomes Using Nanopore Technology
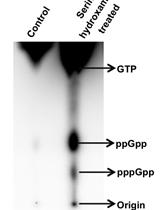
Determination of (p)ppGpp Levels During Stringent Response in Streptomyces coelicolor by Thin Layer Chromatography
Molecular Biology
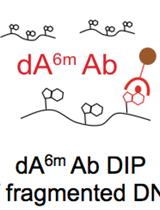
Identification of Methylated Deoxyadenosines in Genomic DNA by dA6m DNA Immunoprecipitation
Neuroscience
Protocol for Primary Microglial Culture Preparation
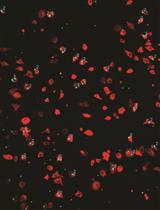
Microglial Phagocytosis Assay
Apparatus and General Methods for Exposing Rats to Audiogenic Stress
Plant Science
Hydrogen Peroxide Measurement in Arabidopsis Root Tissue Using Amplex Red
A Ribosome Footprinting Protocol for Plants
Metabolite Profiling of Mature Arabidopsis thaliana Seeds Using Gas Chromatography-Mass Spectrometry (GC-MS)
Putrescine Biosynthesis Inhibition in Tomato by DFMA and DFMO Treatment
PEA-CLARITY: Three Dimensional (3D) Molecular Imaging of Whole Plant Organs
Determination of Recombinant Mannitol-1-phosphate Dehydrogenase Activity from Ectocarpus sp.
Cytohistological Analyses of Mega-sporogenesis and Gametogenesis in Ovules of Limonium spp.
Stem Cell
Allogeneic Transplantation of Testicular Hyperplasia in rag1 Mutant Zebrafish