- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2025
Volume: 15, Issue: 15
Biochemistry
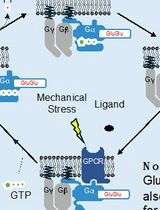
Direct Activity Measurement of Heterotrimeric Gi Proteins and Gq Protein By Effector Pulldown
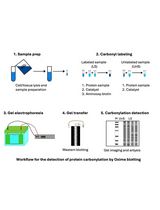
Reliable and Sensitive Detection of Carbonylated Proteins by Oxime Blot
Bioinformatics and Computational Biology
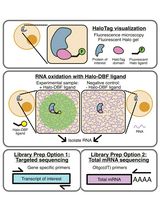
Analyzing RNA Localization Using the RNA Proximity Labeling Method OINC-seq
Cell Biology
Reprogramming White Fat Cells for Adipose Manipulation Transplantation (AMT) Therapy
Accurate Identification of Cell Cycle Stages in RPE1 Cells Using the ImmunoCellCycle-ID Method
Developmental Biology
Fertility test of mice (Mus musculus)
Neuroscience
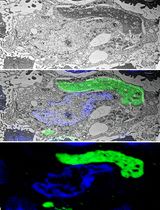
Isolation and Imaging of Microvessels From Brain Tissue
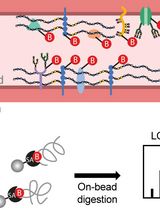
Luminal Cerebrovascular Proteomics
A Step-By-Step Protocol for Correlative Light and Electron Microscopy Imaging of Proteinaceous Deposits in Cultured Cells and Human Brain Tissues
Plant Science
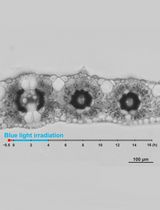
Live Leaf-Section Imaging for Visualizing Intracellular Chloroplast Movement and Analyzing Cell–Cell Interactions
Systems Biology
Protein Turnover Dynamics Analysis With Subcellular Spatial Resolution