- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2024
Volume: 14, Issue: 24
Biophysics
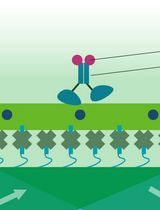
Total Internal Reflection Fluorescence (TIRF) Single-Molecule Assay to Analyze the Motility of Kinesin
Developmental Biology
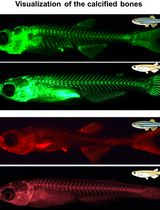
Live Visualization of Calcified Bones in Zebrafish and Medaka Larvae and Juveniles Using Calcein and Alizarin Red S
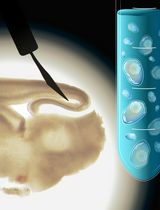
Egg Microinjection for the Ladybird Beetle Harmonia axyridis
Immunology
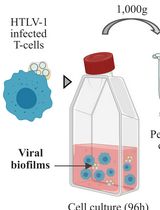
Isolation of Viral Biofilms From HTLV-1 Chronically Infected T Cells and Integrity Analysis
Microbiology
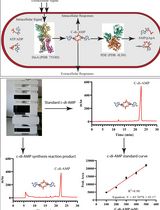
An HPLC-based Assay to Study the Activity of Cyclic Diadenosine Monophosphate (C-di-AMP) Synthase DisA from Mycobacterium smegmatis
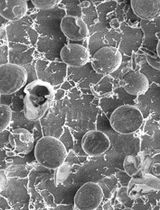
Cryo-SEM Investigation of Chlorella Using Filter Paper as Substrate
Molecular Biology
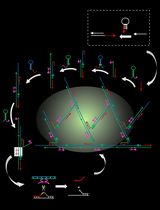
An Autocatalytic Platform Combining a Nonlinear Hybridization Chain Reaction and DNAzyme to Detect microRNA
Assessment of SREBP Activation Using a Microsomal Vesicle Budding Assay
Neuroscience
Microdissection and Single-Cell Suspension of Neocortical Layers From Ferret Brain for Single-Cell Assays
Plant Science
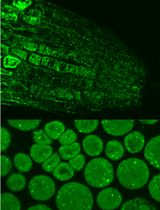
Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Stem Cell
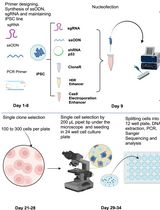
CRISPR/Cas9-Based Protocol for Precise Genome Editing in Induced Pluripotent Stem Cells
Systems Biology
Muscle Biopsy Sample Preparation and Proteomics Analysis Based on UHPLC-MS/MS