- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2016
Volume: 6, Issue: 1
Cancer Biology
PhagoKinetic Track Assay: Imaging and Analysis of Single Cell Migration
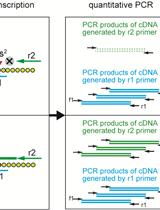
Measurement of 2-methylthio Modifications in Mitochondrial Transfer RNAs by Reverse-transcription Quantitative PCR
Preparation of Recombinant Galectin-3 for Cancer Studies
Immunology
Protocol-In vitro T Cell Proliferation and Treg Suppression Assay with Celltrace Violet
Neuroscience
Fluoro-Jade B Staining for Neuronal Cell Death
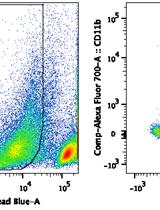
Isolation and Purification of Murine Microglial Cells for Flow Cytometry
Neurite Outgrowth Assay
Plant Science
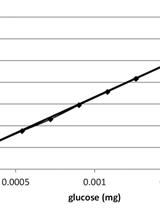
Saccharification Protocol for Small-scale Lignocellulosic Biomass Samples to Test Processing of Cellulose into Glucose
Measurement of Uptake and Root-to-Shoot Distribution of Sulfate in Arabidopsis Seedlings
Super-resolution Imaging of Live BY2 Cells Using 3D-structured Illumination Microscopy
Quantification of Low Molecular Weight Thiols in Arabidopsis