- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2024
Volume: 14, Issue: 14
Biochemistry
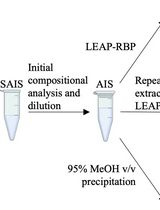
Rapid and Efficient Isolation of Total RNA-Bound Proteomes by Liquid Emulsion–Assisted Purification of RNA-Bound Protein (LEAP-RBP)
Slot Blot Analysis of Intracellular Glyceraldehyde-Derived Advanced Glycation End Products Using a Novel Lysis Buffer and Polyvinylidene Difluoride Membrane
Biophysics
Purification and Cryo-Electron Microscopy Analysis of Bacterial Appendages
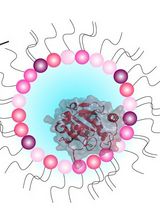
An NMR Approach for Investigating Membrane Protein–Lipid Interactions Using Native Reverse Micelles
Approach for Electrophysiological Studies of Spinal Lamina X Neurons
Cell Biology
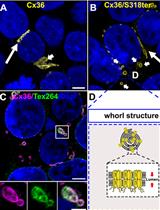
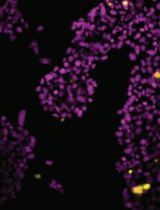
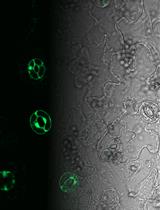
Characterizing ER Retention Defects of PDZ Binding Deficient Cx36 Mutants Using Confocal Microscopy
Molecular Biology
Well Plate–Based Localized Electroporation Workflow for Rapid Optimization of Intracellular Delivery
Plant Science
Analysis of Guard Cell Readouts Using Arabidopsis thaliana Isolated Epidermal Peels