- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2024
Volume: 14, Issue: 11
Cancer Biology
A New Approach for Assessment of Neutrophil Extracellular Traps Through Immunofluorescence Staining in Whole Blood Smears
Cell Biology
Isolation and Characterization of Extracellular Vesicles Derived from Ex Vivo Culture of Visceral Adipose Tissue
Environmental science
Rearing and Shipping of Uranotaenia lowii, a Frog-Biting Mosquito
Immunology
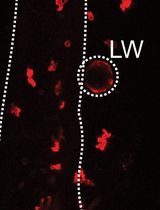
Visualising Neutrophil Actin Dynamics in Zebrafish in Response to Laser Wounding Using Two-Photon Microscopy
Microbiology
A Multi-Color Immunofluorescence Assay to Distinguish Intracellular From External Leishmania Parasites
Molecular Biology
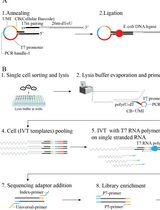
Linearly Amplified Single-Stranded RNA-Derived Transcriptome Sequencing (LAST-seq)
Plant Science
CRISPR-Cas9 Protocol for Efficient Gene Knockout and Transgene-free Plant Generation
Fast, Easy, and Comprehensive Techniques for Microscopic Observations of Fungal and Oomycete Organisms Inside the Roots of Herbaceous and Woody Plants