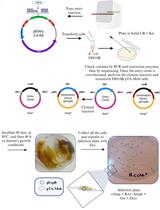
Analysis of Cleavage Activity of Dengue Virus Protease by Co-transfections
The genome of the dengue virus codes for a single polypeptide that yields three structural and seven non-structural (NS) proteins upon post-translational modifications. Among them, NS protein-3 (NS3) possesses protease activity, involved in the processing of the self-polypeptide and in the cleavage of host proteins. Identification and analysis of such host proteins as substrates of this protease facilitate the development of specific drugs. In vitro cleavage analysis has been applied, which requires homogeneously purified components. However, the expression and purification of both S3 and erythroid differentiation regulatory factor 1 (EDRF1) are difficult and unsuccessful on many occasions. EDRF1 was identified as an interacting protein of dengue virus protease (NS3). The amino acid sequence analysis indicates the presence of NS3 cleavage sites in this protein. As EDRF1 is a high-molecular-weight (~138 kDa) protein, it is difficult to express and purify the complete protein. In this protocol, we clone the domain of the EDRF1 protein (C-terminal end) containing the cleavage site and the NS3 into two different eukaryotic expression vectors containing different tags. These recombinant vectors are co-transfected into mammalian cells. The cell lysate is subjected to SDS-PAGE followed by western blotting with anti-tag antibodies. Data suggest the disappearance of the EDRF1 band in the lane co-transfected along with NS3 protease but present in the lane transfected with only EDRF1, suggesting EDRF1 as a novel substrate of NS3 protease. This protocol is useful in identifying the substrates of viral-encoded proteases using ex vivo conditions. Further, this protocol can be used to screen anti-protease molecules.Key features• This protocol requires the cloning of protease and substrate into two different eukaryotic expression vectors with different tags.• Involves the transfection and co-transfection of both the above recombinant vectors individually and together.• Involves western blotting of the same PVDF membrane containing total proteins of the cell lysate with two different antibodies.• Does not require purified proteins for the analysis of cleavage of any suspected substrate by the protease.Graphical overview