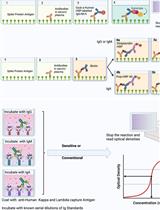
An Optimised Indirect ELISA Protocol for Detection and Quantification of Anti-viral Antibodies in Human Plasma or Serum: A Case Study Using SARS-CoV-2
Advanced immunoassays are crucial in assessing antibody responses, serving immune surveillance goals, characterising immunological responses to evolving viral variants, and guiding subsequent vaccination initiatives. This protocol outlines an indirect ELISA protocol to detect and quantify virus-specific antibodies in plasma or serum after exposure to viral antigens. The assay enables the measurement of IgG, IgA, and IgM antibodies specific to the virus of interest, providing qualitative and quantitative optical densities and concentration data. Although this protocol refers to SARS-CoV-2, its methodology is versatile and can be modified to assess antibody responses for various viral infections and to evaluate vaccine trial outcomes.Key features• This protocol builds upon previously described methodology [1] explicitly tailored for SARS-CoV-2 and broadens its applicability to other viral infections.• The protocol outlines establishing antibody responses to SARS-CoV-2 infections by determining optical densities and concentrations from blood plasma or serum.Graphical overview Summary of the conventional ELISA (A) vs. sensitive ELISA (B) procedures. In both A and B, wells are coated with a capture antigen, such as the spike protein, while in (C) they are coated with human Kappa and Lambda capture antibodies. For the conventional ELISA (A), wells with immobilised capture antigens receive serum/plasma containing the target antibody (A1 and B1). This is followed by an HRP-conjugated detection antibody specific to the captured antibody (A2 and B2) and then a substrate solution that reacts with the HRP, producing a colour proportional to the concentration of the antibody in the serum/plasma (A3 and B3). The reaction is halted, and absorbance is measured. In the sensitive ELISA (B), after the serum/plasma addition (A1 and B1), a Biotin-conjugated primary detection antibody is introduced (A2 and B2). Depending on the target antibody, a secondary streptavidin-HRP conjugated detection antibody is added for IgG or IgM (3a) or a poly-HRP 40 detection antibody for IgA (3b). A substrate is introduced, producing a colour change proportional to the antibody concentration (A4 and B4). The reaction is then stopped, and absorbance is measured. In Panel C, wells are coated with human Kappa and Lambda capture antibodies. Serial dilutions of a known antibody standard are introduced. After undergoing the standard ELISA steps, a detection antibody is added, specifically binding to the Ig standard antibody. Subsequently, a substrate solution causes a colour change proportional to the antibody concentration in the serum/plasma. The reaction is halted, and the absorbance of each well is measured. The resulting optical densities from the coated wells form the standard curve, plotting the absorbance against concentrations.