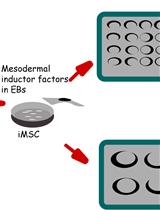
Efficient Differentiation of Human Induced Pluripotent Stem Cell (hiPSC)-Derived Mesenchymal Progenitors Into Adipocytes and Osteoblasts
Human induced pluripotent stem cells (hiPSCs) hold immense promise in regenerative medicine as they can differentiate into various cell lineages, including adipocytes, osteoblasts, and chondrocytes. Precisely guiding hiPSC-derived mesenchymal progenitor cells (iMSCs) towards specific differentiation pathways is crucial for harnessing their therapeutic potential in tissue engineering, disease modeling, and regenerative therapies. To achieve this, we present a comprehensive and reproducible protocol for effectively differentiating iMSCs into adipocytes and osteoblasts. The differentiation process entails culturing iMSCs in tailored media supplemented with specific growth factors, which act as cues to initiate adipogenic or osteogenic commitment. Our protocol provides step-by-step guidelines for achieving adipocyte and osteoblast differentiation, ensuring the generation of mature and functional cells. To validate the success of differentiation, key assessment criteria are employed. For adipogenesis, the presence of characteristic lipid droplets within the iMSC-derived cells is considered indicative of successful differentiation. Meanwhile, Alizarin Red staining serves as a marker for the osteogenic differentiation, confirming the formation of mineralized nodules. Importantly, the described method stands out due to its simplicity, eliminating the need for specialized equipment, expensive materials, or complex reagents. Its ease of implementation offers an attractive advantage for researchers seeking robust and cost-effective approaches to derive adipocytes and osteoblasts from iMSCs. Overall, this protocol establishes a foundation for exploring the therapeutic potential of hiPSC-derived cells and advancing the field of regenerative medicine.Key features• iMSC derivation in this protocol uses embryonic body formation technique.• Adipogenesis and osteogenesis protocols were optimized for human iPSC-derived iMSCs.• Derivation of iMSC from hiPSC was developed in a feeder-free culture condition.• This protocol does not include human iPSC reprogramming strategies.Graphical overviewSchematic representation of induced pluripotent stem cell (iPSC) differentiation into adipocytes and osteoblasts via mesenchymal progenitors as intermediates