- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2023
Volume: 13, Issue: 16
Bioinformatics and Computational Biology
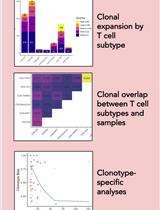
T Cell Clonal Analysis Using Single-cell RNA Sequencing and Reference Maps
Biophysics
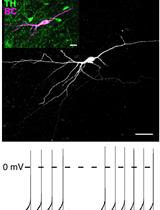
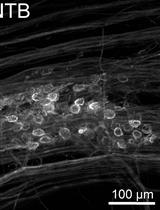
Perforated Patch Clamp Recordings in ex vivo Brain Slices from Adult Mice
Cancer Biology
Quantification of Chromosomal Aberrations in Mammalian Cells
Cell Biology
Stereotactic Delivery of Helper-dependent Adenoviral Viral Vectors at Distinct Developmental Time Points to Perform Age-dependent Molecular Manipulations of the Mouse Calyx of Held
Medicine
Catheterization of Pulmonary and Carotid Arteries for Concurrent Measurement of Mean Pulmonary and Systemic Arterial Pressure in Rat Models of Pulmonary Arterial Hypertension
Microbiology
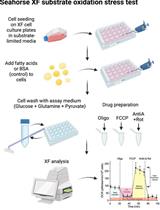
Mass Spectrometry-based Lipidomics, Lipid Bioenergetics, and Web Tool for Lipid Profiling and Quantification in Human Cells
Determination of Poly(3-hydroxybutyrate) Content in Cyanobacterium Synechocystis sp. PCC 6803 Using Acid Hydrolysis Followed by High-performance Liquid Chromatography
Molecular Biology
In Vitro Analysis of Stalled Ribosomes using Puromycin Incorporation
Neuroscience
Multi-color Flow Cytometry Protocol to Characterize Myeloid Cells in Mouse Retina Research
Detection and Quantification of Calcium Ions in the Endoplasmic Reticulum and Cytoplasm of Cultured Cells Using Fluorescent Reporter Proteins and ImageJ Software
Caste Transition and Reversion in Harpegnathos saltator Ant Colonies
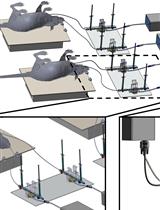
A Method for Studying Social Signal Learning of the Waggle Dance in Honey Bees
Plant Science
Fluorescent Biosensor Imaging of Nitrate in Arabidopsis thaliana
Quantification of Botrytis cinerea Growth in Arabidopsis thaliana
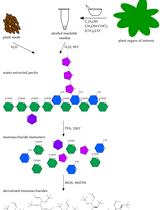
Analysis of Pectin-derived Monosaccharides from Arabidopsis Using GC–MS
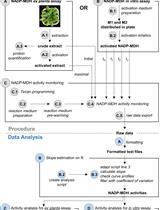
A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
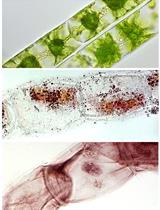
Improved Methods for Acetocarmine and Haematoxylin Staining to Visualize Chromosomes in the Filamentous Green Alga Zygnema (Charophyta)
Stem Cell
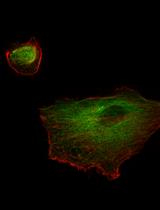
Preparation of Human Kidney Progenitor Cultures and Their Differentiation into Podocytes