- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2023
Volume: 13, Issue: 15
Biochemistry
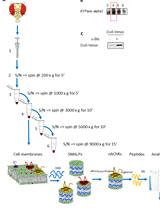
Enrichment of Membrane Proteins for Downstream Analysis Using Styrene Maleic Acid Lipid Particles (SMALPs) Extraction
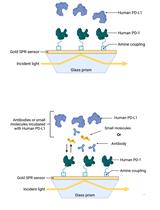
Establishment of Human PD-1/PD-L1 Blockade Assay Based on Surface Plasmon Resonance (SPR) Biosensor
Cancer Biology
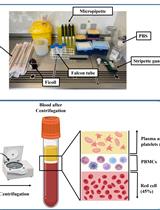
Ex vivo Drug Sensitivity Imaging-based Platform for Primary Acute Lymphoblastic Leukemia Cells
Cell Biology
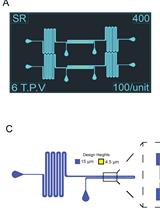
Fabrication of Microfluidic Devices for Continuously Monitoring Yeast Aging
VAR2CSA Ectodomain Labeling in Plasmodium falciparum Infected Red Blood Cells and Analysis via Flow Cytometry
Developmental Biology
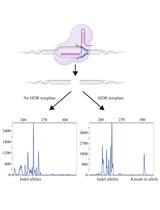
Fluorescent PCR–based Screening Methods for Precise Knock-in of Small DNA Fragments and Point Mutations in Zebrafish
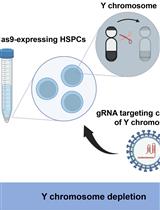
Development of a Mouse Model of Hematopoietic Loss of Y Chromosome
Immunology
Label-free Chemical Characterization of Polarized Immune Cells in vitro and Host Response to Implanted Bio-instructive Polymers in vivo Using 3D OrbiSIMS
Microbiology
Protocol for the High-quality Plasmid Isolation from Different Recalcitrant Bacterial Species: Agrobacterium spp., Rhizobium sp., and Bacillus thuringiensis
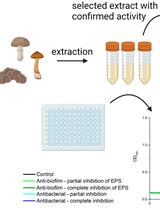
SIMBA Method—Simultaneous Detection of Antimicrobial and Anti-biofilm Activity of New Compounds Using Salmonella Infantis
Molecular Biology
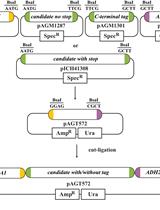
A qPCR Method to Distinguish between Expression of Transgenic and Endogenous Copies of Genes
Neuroscience
Using Fiber Photometry in Mice to Estimate Fluorescent Biosensor Levels During Sleep
Construction of Activity-based Anorexia Mouse Models
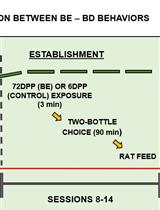
Binging from Food to Alcohol: A Sequential Interaction Between Binging Behaviors in Male Wistar Rats
Plant Science
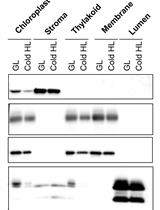
A Simple Sonication Method to Isolate the Chloroplast Lumen in Arabidopsis thaliana
Simple Growth Complementation Assay in Yeast
Bi-directional Dual-flow-RootChip for Physiological Analysis of Plant Primary Roots Under Asymmetric Perfusion of Stress Treatments
Engineering Agrobacterium tumefaciens with a Type III Secretion System to Express Type III Effectors
A Novel Method for Measuring Mitochondrial Respiratory Parameters in Wheat Paleae (Paleae Superior) Using the XF24 Analyzer