- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2023
Volume: 13, Issue: 2
Biophysics
Lysate-to-grid: Rapid Isolation of Native Complexes from Budding Yeast for Cryo-EM Imaging
Developmental Biology
A Reliable and Consistent Method to Quantify Percent Lethality and Life Span in Drosophila melanogaster
Drug Discovery
Methods to Detect AUTOphagy-Targeting Chimera (AUTOTAC)-mediated Targeted Protein Degradation in Tauopathies
Molecular Biology
Preparation and Characterization of DNA-assembled GRS-DNA-CuS Nanodandelions
Neuroscience
Establishment of Restraint Stress–induced Anorexia and Social Isolation–induced Anorexia Mouse Models
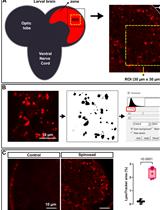
In vivo Assessment of Lysosomal Stress in the Drosophila Brain Using Confocal Fluorescence Microscopy
Plant Science
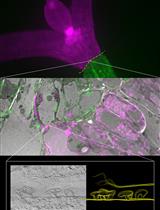
Targeting Ultrastructural Events at the Graft Interface of Arabidopsis thaliana by A Correlative Light Electron Microscopy Approach
Evaluating Plant Drought Resistance with a Raspberry Pi and Time-lapse Photography
Stem Cell
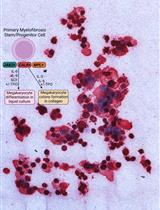
Thrombopoietin-independent Megakaryocyte Differentiation of Hematopoietic Progenitor Cells from Patients with Myeloproliferative Neoplasms
Systems Biology
Rapid Multiplexed Flow Cytometric Validation of CRISPRi sgRNAs in Tissue Culture