- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2022
Volume: 12, Issue: 23
Biochemistry
Assessing the in vitro Binding Affinity of Protein–RNA Interactions Using an RNA Pull-down Technique
Cancer Biology
Fluorescence Time-lapse Imaging of Entosis Using Tetramethylrhodamine Methyl Ester Staining
Immunology
Improved Macrophage Enrichment from Mouse Skeletal Muscle
Human Auto-IgG Purification from High Volume Serum Sample by Protein G Affinity Purification
Medicine
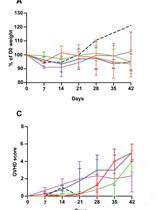
Modelling Graft-Versus-Host Disease in Mice Using Human Peripheral Blood Mononuclear Cells
Molecular Biology
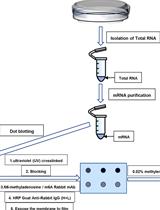
Analysis of N6-methyladenosine RNA Modification Levels by Dot Blotting
Neuroscience
Infection of the Developing Central Nervous System of Drosophila by Mammalian Eukaryotic and Prokaryotic Pathogens
Conditioned Lick Suppression: Assessing Contextual, Cued, and Context-cue Compound Fear Responses Independently of Locomotor Activity in Mice
Plant Science
Focused Ion Beam Milling and Cryo-electron Tomography Methods to Study the Structure of the Primary Cell Wall in Allium cepa
Measurement of Transgenes Copy Number in Wheat Plants Using Droplet Digital PCR