- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2022
Volume: 12, Issue: 21
Biological Engineering
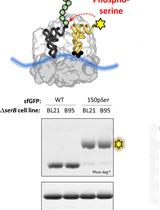
Site-specific Incorporation of Phosphoserine into Recombinant Proteins in Escherichia coli
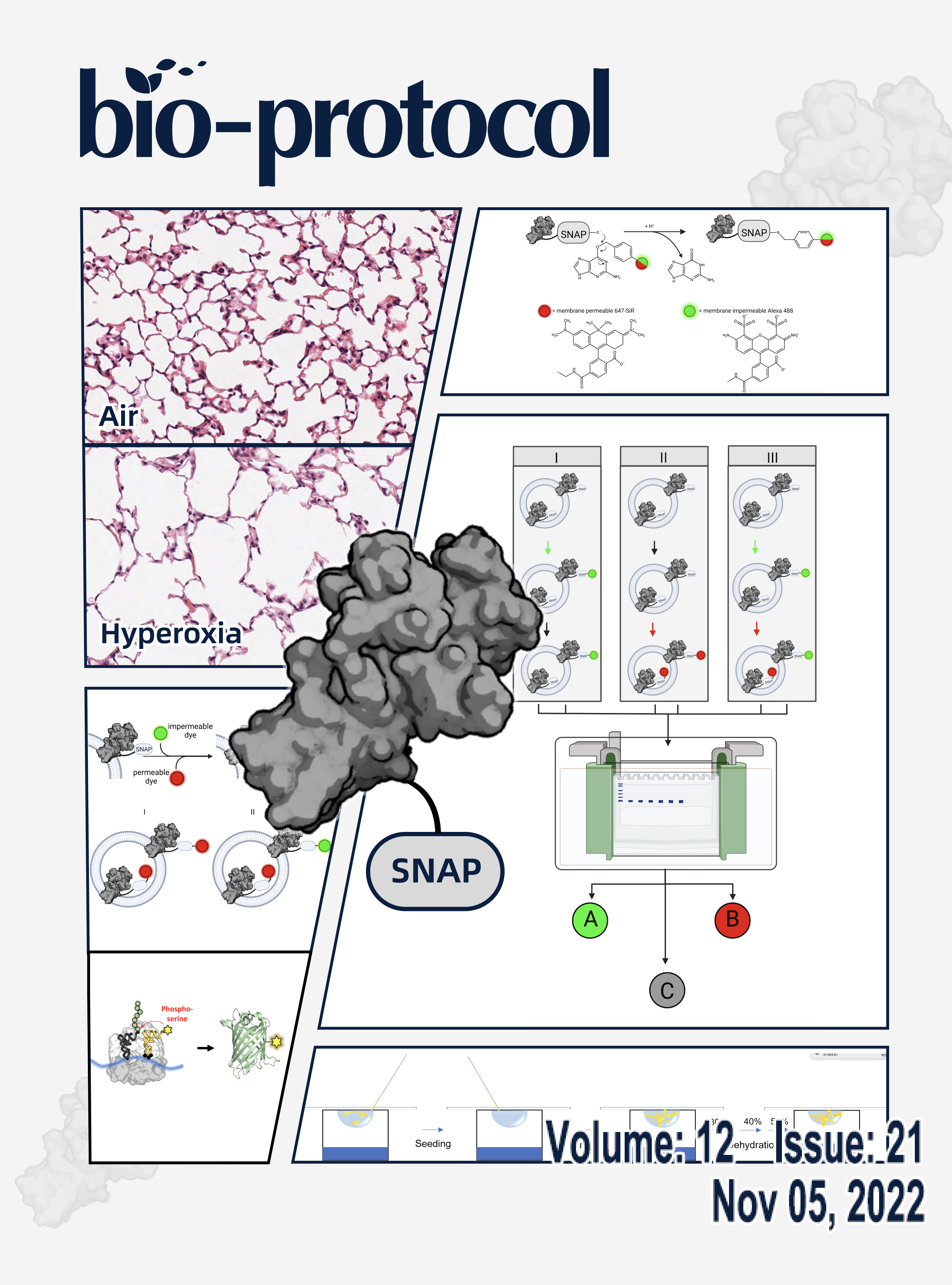
A Fluorescence-based Approach Utilizing Self-labeling Enzyme Tags to Determine Protein Orientation in Large Unilamellar Vesicles
Biophysics
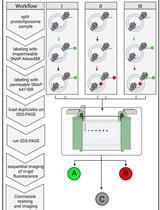
In situ cryo-FIB/SEM Specimen Preparation Using the Waffle Method
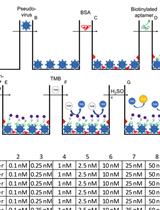
Binding Affinity Measurements Between DNA Aptamers and their Virus Targets Using ELONA and MST
Cancer Biology
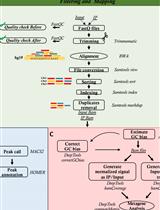
OxiDIP-Seq for Genome-wide Mapping of Damaged DNA Containing 8-Oxo-2'-Deoxyguanosine
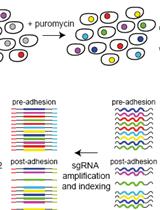
An Unbiased CRISPR-Cas9 Screening Method for the Identification of Positive and Negative Regulatory Proteins of Cell Adhesion
Immunology
Murine Double Hit Model for Neonatal Cardiopulmonary Diseases: Bronchopulmonary Dysplasia (BPD) and Pulmonary Hypertension Associated with BPD
Mechanobiology
Motion-capture Analysis of Mice Using a Video Recorded on an iPhone Camera
Microbiology
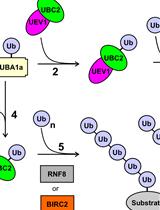
In vitro Di-ubiquitin Formation Assay and E3 Cooperation Assay
Plant Science
X-ray Crystallography: Seeding Technique with Cytochrome P450 Reductase