- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2022
Volume: 12, Issue: 13
Biochemistry
Whole-mount Senescence-Associated Beta-Galactosidase (SA-β-GAL) Activity Detection Protocol for Adult Zebrafish
Biological Engineering
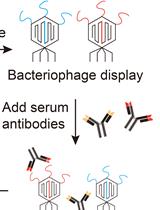
VirScan: High-throughput Profiling of Antiviral Antibody Epitopes
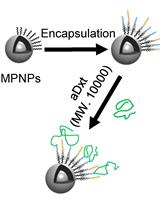
A Robust Nanoparticle-based Magnetic Separation Method for Intact Lysosomes
Cell Biology
Experimental Models for Cold Exposure of Muscle in vitro and in vivo
Developmental Biology
In vivo Characterization of Endogenous Protein Interactomes in Drosophila Larval Brain, Using a CRISPR/Cas9-based Strategy and BioID-based Proximity Labeling
Immunology
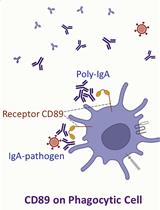
Serological Measurement of Poly-IgA Immune Complex Levels in IgA Nephropathy and IgA Vasculitis
Microbiology
A β-glucuronidase (GUS) Based Bacterial Competition Assay to Assess Fine Differences in Fitness during Plant Infection
Molecular Biology
HaloChIP-seq for Antibody-Independent Mapping of Mouse Transcription Factor Cistromes in vivo
Neuroscience
Automated Quantification of Multiple Cell Types in Fluorescently Labeled Whole Mouse Brain Sections Using QuPath