- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2015
Volume: 5, Issue: 17
Biochemistry
Immunolocalization of Proteins in Corals: the V-type H+-ATPase Proton Pump
Post-crystallization Improvement of RNA Crystals by Synergistic Ion Exchange and Dehydration
Cancer Biology
[14C]-Tryptophan Metabolic Tracing in Liver Cancer Cells
Immunology
Thioglycollate-elicited Peritoneal Macrophages Preparation and Arginase Activity Measurement in IL-4 Stimulated Macrophages
Chitin-challenged Mice Model to Study M2 Macrophages Polarization
Microbiology
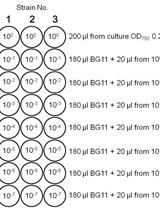
Spot Assays for Viability Analysis of Cyanobacteria
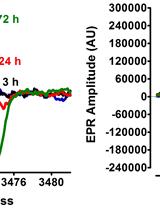
Electron Paramagnetic Resonance (EPR) Spectroscopy to Detect Reactive Oxygen Species in Staphylococcus aureus
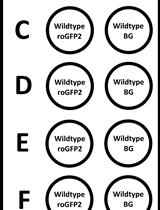
In vitro Real-time Measurement of the Intra-bacterial Redox Potential
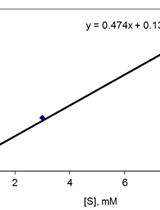
Determination of Quinone Reductase Activity
Determination of Hydroquinone Dioxygenase Activity
Plant Science
Cryo-focused Ion Beam Sample Preparation for Imaging Vitreous Cells by Cryo-electron Tomography
Expression and Partial Purification of His-tagged Proteins in a Plant System
A Bioimaging Pipeline to Show Membrane Trafficking Regulators Localized to the Golgi Apparatus and Other Organelles in Plant Cells
Detection of Poly (A) RNA in Mesophyll Cells of Nicotiana benthamiana Using in situ Hybridization
A Non-Radioactive Method for Measuring PP2A Activity in Plants







![[14C]-Tryptophan Metabolic Tracing in Liver Cancer Cells](https://en-cdn.bio-protocol.org/imageup/arcimg/20150906122132762.jpg?t=1770108251)