- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2021
Volume: 11, Issue: 23
Biological Engineering
A low-cost Portable Device to Deliver Smoke, Volatile or Vaporized Substances to Drosophila melanogaster , Useful for Research and/or Educational Assays
Cell Biology
Quantitative Determination of Primary Cilia Protein Distribution Using Immunofluorescence Staining and MATLAB Analysis
Isolation of Healthy F4/80+ Macrophages from Embryonic day E13.5 Mouse Fetal Liver Using Magnetic Nanoparticles for Single Cell Sequencing
In vitro Fluid Shear Stress Induced Sclerostin Degradation and CaMKII Activation in Osteocytes
A Simple Method for in situ Quantification of Cells on Carriers
Developmental Biology
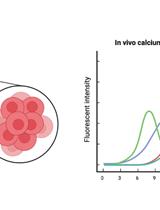
In vivo Imaging of Calcium Activities from Pancreatic β-cells in Zebrafish Embryos Using Spinning-disc Confocal and Two-photon Light-sheet Microscopy
Ex-vivo Microtubule Stability Assay Using Drosophila Wing Disc
Immunology
Bacterial Infection with Listeria monocytogenes in Mice and Subsequent Analysis of Antigen-Specific CD8 T Cell Responses
Microbiology
A Phenotypic Screen for the Liver Stages of Plasmodium vivax
A Retro-orbital Sinus Injection Mouse Model to Study Early Events and Reorganization of the Astrocytic Network during Pneumococcal Meningitis
Neuroscience
Immunoprecipitation for Protein-Protein Interactions and for RNA Enrichment in Drosophila melanogaster
Conditional Gene Editing in Presynaptic Extinction-ensemble Cells via the CRISPR-SaCas9 System
Plant Science
Split-luciferase Complementation Imaging Assay to Study Protein-protein Interactions in Nicotiana benthamiana
Isolation of Plant Nuclei Compatible with Microfluidic Single-nucleus ATAC-sequencing
Stem Cell
A Simple Method for the Isolation and in vitro Expansion of Highly Pure Mouse and Human Satellite Cells
Systems Biology
TGIRT-seq Protocol for the Comprehensive Profiling of Coding and Non-coding RNA Biotypes in Cellular, Extracellular Vesicle, and Plasma RNAs