- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2021
Volume: 11, Issue: 10
Biochemistry
A Fluorescence Dequenching-based Liposome Leakage Assay to Measure Membrane Permeabilization by Pore-forming Proteins
Intracellular IRF5 Dimerization Assay
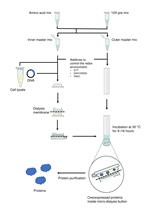
Cell-free Synthesis of Correctly Folded Proteins with Multiple Disulphide Bonds: Production of Fungal Hydrophobins
Biophysics
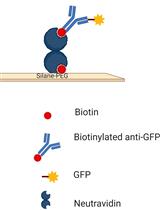
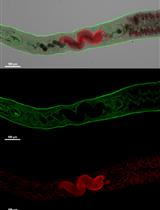
Single-Molecule Studies of Membrane Receptors from Brain Region Specific Nanovesicles
Cancer Biology
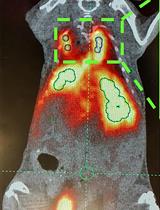
Surface Engineering and Multimodal Imaging of Multistage Delivery Vectors in Metastatic Breast Cancer
Cell Biology
FACS Enrichment of Total Interstitial Cells and Fibroblasts from Adult Mouse Ventricles
Developmental Biology
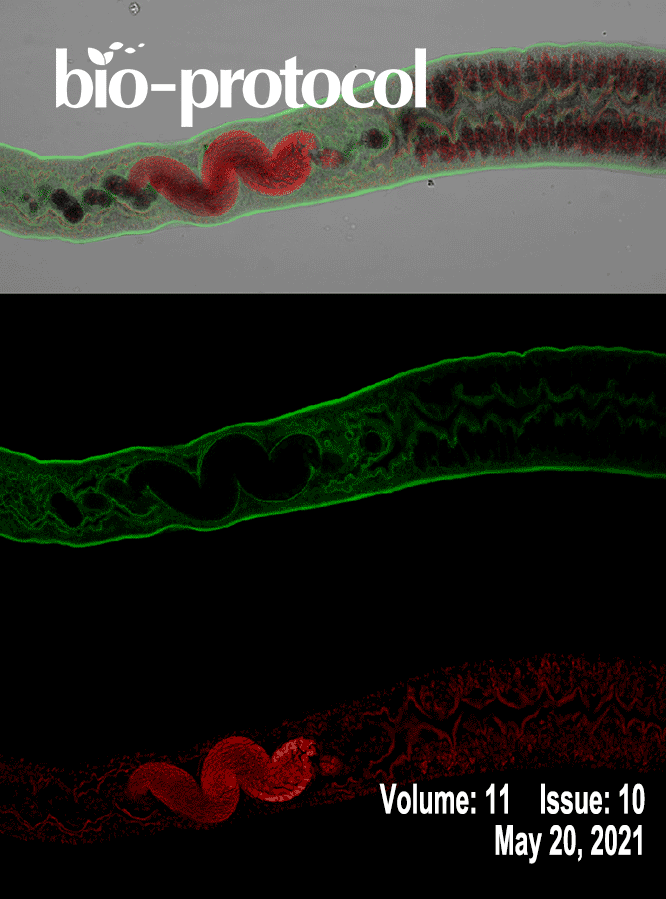
Imaging and Fluorescence Quantification in Caenorhabditis elegans with Flow Vermimetry and Automated Microscopy
Immunology
In vitro and In vivo CD8+ T Cell Suppression Assays
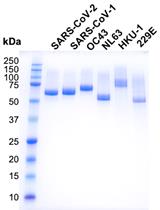
Production of the Receptor-binding Domain of the Viral Spike Proteins from 2003 and 2019 SARS CoVs and the Four Common Human Coronaviruses for Serologic Assays and Inhibitor Screening
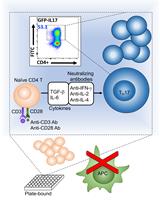
A Simple and Robust Protocol for in vitro Differentiation of Mouse Non-pathogenic T Helper 17 Cells from CD4+ T Cells
Microbiology
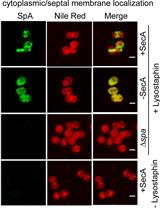
Tracking the Subcellular Localization of Surface Proteins in Staphylococcus aureus by Immunofluorescence Microscopy
ODELAM: Rapid Sequence-independent Detection of Drug Resistance in Mycobacterium tuberculosis Isolates
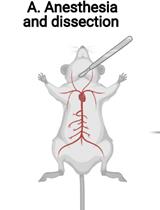
Parasitemia Evaluation in Mice Infected with Schistosoma mansoni
Molecular Biology
A New Method for Studying RNA-binding Proteins on Specific RNAs
In vivo CD40 Silencing by siRNA Infusion in Rodents and Evaluation by Kidney Immunostaining
Neuroscience
Operant Vapor Self-administration in Mice
Stem Cell
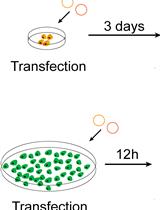
Ex vivo Tissue Culture Protocols for Studying the Developing Neocortex