- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2015
Volume: 5, Issue: 15
Cancer Biology
DNA in situ Hybridizations for VEGFA Gene Locus (6p12) in Human Tumor Tissue
Cell Biology
Assessing Mitochondrial Transport via Cytoplasmic Nanotubular Bridges in Cells
Developmental Biology
Mouse Retinal Whole Mounts and Quantification of Vasculature Protocol
Immunology
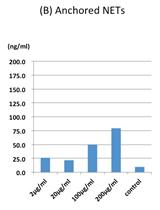
Quantification of ex vivo Neutrophil Extracellular Traps
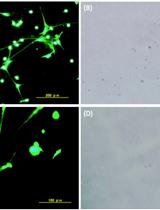
Visualization of ex vivo Neutrophil Extracellular Traps by Fluorescence Microscopy
Microbiology
XTT Assay of Antifungal Activity
Evaluation of Nodulation Speed by Sinorhizobium Strains
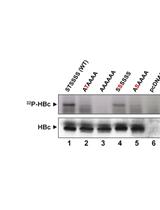
Detection of HBV C Protein Phosphorylation in the Cell
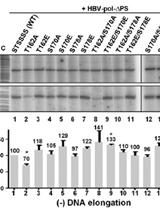
Primer Extension Analysis of HBV DNA with Strand-Specific Primers
Quantitative Evaluation of Competitive Nodulation among Different Sinorhizobium Strains
Neuroscience
Measuring Blood-brain-barrier Permeability Using Evans Blue in Mice
Plant Science
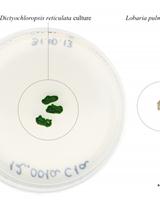
Genomic DNA Extraction and Genotyping of Dictyochloropsis Green Algae Strains
Tissue Culturing and Harvesting of Protonemata from the Moss Physcomitrella patens
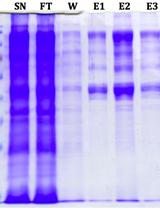
Expression and Purification of the Arabidopsis E4 SUMO Ligases PIAL1 and PIAL2
Systems Biology
Protocol for the Generation of a Transcription Factor Open Reading Frame Collection (TFome)